Hiii, thanks for the reply! (sorry I went to a department retreat in the past few days)
I tried the following steps:
3dcopy mydata.nii.gz mydata
3dinfo mydata+orig
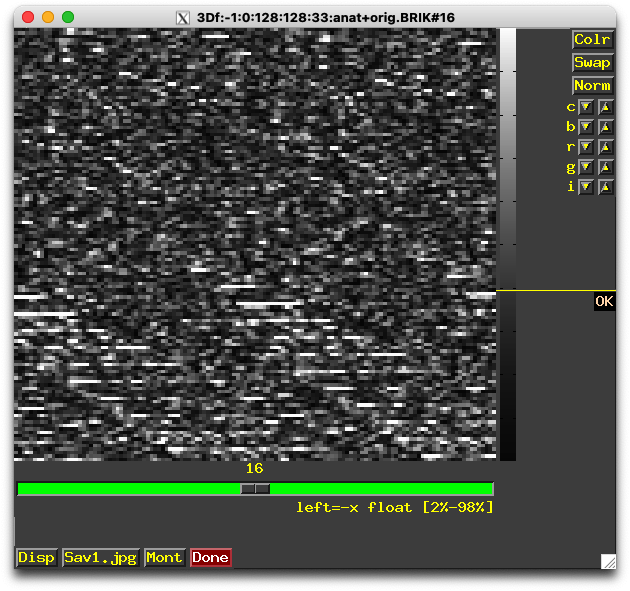
to3d ‘3D:-1:0:128:128:33:mydata+orig.BRIK’
3dcopy output:
++ 3dcopy: AFNI version=AFNI_24.1.16 (Jun 6 2024) [64-bit]
** AFNI converts NIFTI_datatype=64 (FLOAT64) in file /Users/yibeichen/Desktop/fusi/fusi.nii to FLOAT32
Warnings of this type will be muted for this session.
Set AFNI_NIFTI_TYPE_WARN to YES to see them all, NO to see none.
*+ WARNING: NO spatial transform (neither qform nor sform), in NIfTI file '/Users/yibeichen/Desktop/fusi/fusi.nii'
3dinfo output:
++ 3dinfo: AFNI version=AFNI_24.1.16 (Jun 6 2024) [64-bit]
Dataset File: fusi+orig
Identifier Code: AFN_yLaLnmhciQBuIYQCRewhJQ Creation Date: Wed Jun 12 12:57:11 2024
Template Space: ORIG
Dataset Type: Anat Bucket (-abuc)
Byte Order: LSB_FIRST [this CPU native = LSB_FIRST]
Storage Mode: BRIK
Storage Space: 20,062,464 (20 million) bytes
Geometry String: "MATRIX(-1,0,0,0,0,-1,0,0,0,0,1,0):192,151,173"
Data Axes Tilt: Plumb
Data Axes Orientation:
first (x) = Left-to-Right
second (y) = Posterior-to-Anterior
third (z) = Inferior-to-Superior [-orient LPI]
R-to-L extent: -191.000 [R] -to- 0.000 -step- 1.000 mm [192 voxels]
A-to-P extent: -150.000 [A] -to- 0.000 -step- 1.000 mm [151 voxels]
I-to-S extent: 0.000 -to- 172.000 [S] -step- 1.000 mm [173 voxels]
Number of values stored at each pixel = 1
-- At sub-brick #0 '#0' datum type is float: 4.50397 to 83020.4
to3d messages/warnings:
*+ WARNING: *** ILLEGAL INPUTS (cannot save) ***
Axes orientations are not consistent!
++ Making widgets++
++
Hints disabled: X11 failure to create LiteClue window
++
.....
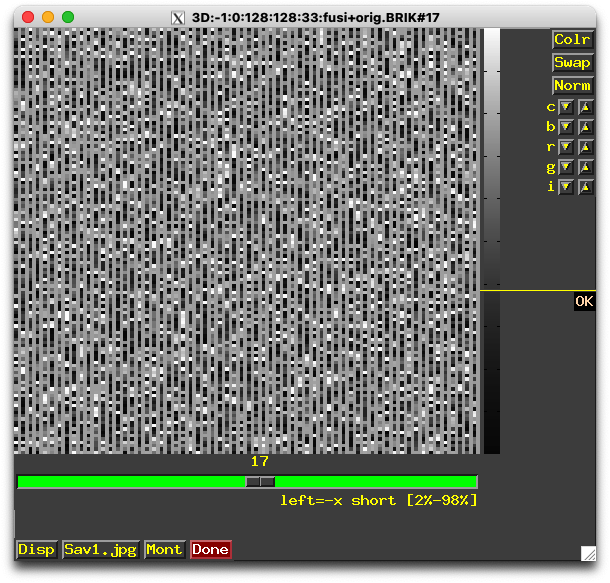
yibeichen@dhcp-10-29-164-213 fusi % to3d '3D:-1:0:128:128:33:fusi+orig.BRIK'
++ to3d: AFNI version=AFNI_24.1.16 (Jun 6 2024) [64-bit]
++ Authored by: RW Cox
++ It is best to use to3d via the Dimon program.
++ Counting images: total=33 2D slices
++ Each 2D slice is 128 X 128 pixels
++ Image data type = short
++ Reading images: .................................
++ to3d WARNING: 134870 negative voxels (24.9449%) were read in images of shorts.
++ It is possible the input images need byte-swapping.
Consider also -ushort2float.
*+ WARNING: *** ILLEGAL INPUTS (cannot save) ***
Axes orientations are not consistent!
++ Making widgets++
++
Hints disabled: X11 failure to create LiteClue window
++
I guess this one is important?
134870 negative voxels (24.9449%) were read in images of shorts.
It is possible the input images need byte-swapping.
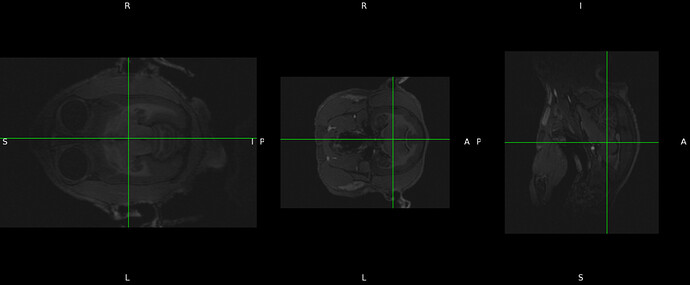
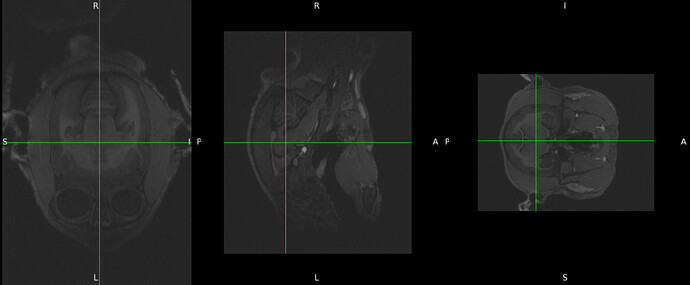
then if I click “view images” I got the following:
I also tried MRIcroGL from here. for some reason, I couldn’t get the correct results after rotation…