Hi, I’m new to AFNI, when I was using AFNI to preprocess dog fMRI images, something goes wrong. I used uber_subject.py to generate the preprocessing scripts. my processing blocks contain tshift align volreg blur mask scale regress. But the images after align is completely distorted. I found that the align block uses 3dSkullStrip to anat image, and this function only apply to human brains. so how should I do?
I should mention that uber_subject.py hasn’t been updated to keep pace with afni_proc.py (the underlying script that uber_subject) calls. I would recommend that you use afni_proc.py directly with various options for considerably more flexibility!
Either way, if you want turn off the skull-stripping in afni_proc.py, pass the option -anat_has_skull no.
Thank you, I will try this
Additional questions, the template for dog brain I used do not have skull, so if I want to strip the skull for the dog brains, how can I do, the 3dSkullStrip do not apply to animals.
Howdy-
I have done a small amount of canine MRI processing previously, and I think we can try using AFNI’s @animal_warper program for this, and integrate the results into a afni_proc.py processing stream. Are you able to share the data, which might make it easier to get started?
Also, for AFNI-specific questions, using this AFNI Message Board link might be easier:
https://afni.nimh.nih.gov/afni/community/board/list.php?1
or, if asking questions here about AFNI programs, please use the “afni” tag. That will make it easier for myself and others to get the notifications (I am not sure how to get notifications about non-tagged keywords on this message board, unfortunately).
–pt
Thank you for your answer! I found this program and I’m trying it out. And, there is one sample data for you(https://drive.google.com/drive/folders/1ktpC1JkE6HTLlX0-Ems8T0EHB9hciXJi?usp=sharing), including 4 epi run, anat, and stimuli onset txt file, and template for tlrc (if it is needed. I’m not sure). If it’s convenient, could you share your processing code for this sample data?
Yeah, you give a good suggestion for the question-asking rules. Thanks again.
Hi-
Yes, I just requested access.
I have gotten a publicly available canine template and tried to fix a number of properties in it… I will try to use that for processing and will be able to share the script, yes.
–pt
Thanks very much! I have tried a lot but there’s always something wrong. ps. I attended AFNI bootcamp in Shenzhen, so we have met before
Ah, cool, then yes, we probably have met. That Bootcamp seems a looong time ago now.
–pt
Hi-
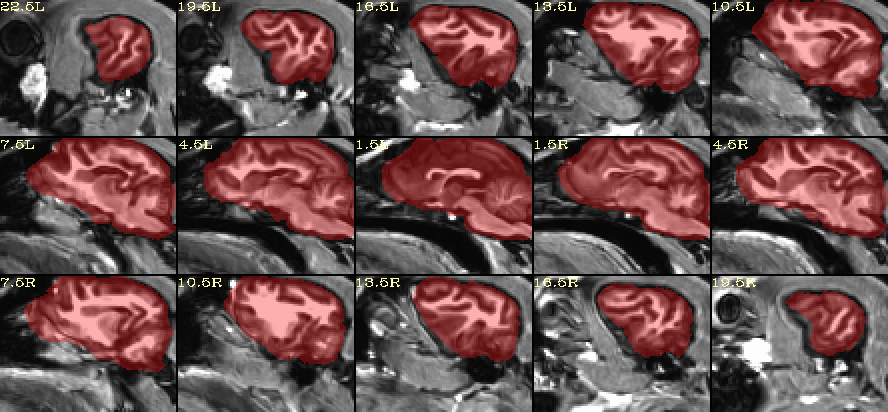
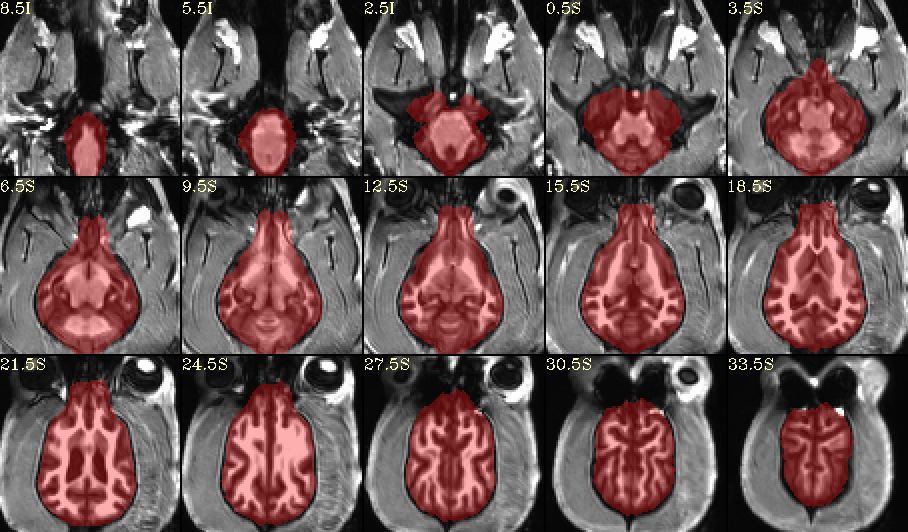
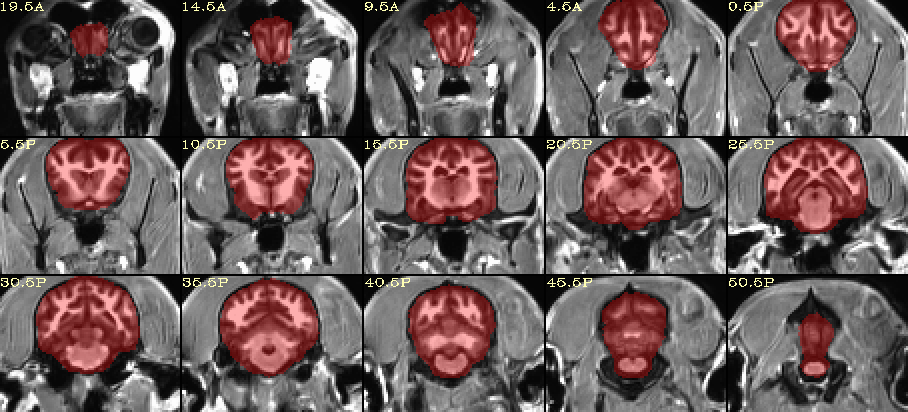
How does this look for skullstripping?
I created this using AFNI’s @animal_warper program, and a modified version of the Johnson & Barry ( 2019) canine template:
Johnson, Philippa J., and Erica F. Barry. (2019) Cortical Atlas of the Canine Brain [dataset]. Cornell University Library eCommons Repository. https://doi.org/10.7298/4t8z-aw34.
Cortical Atlas of the Canine Brain
Note the @animal_warper program also calculates a nonlinear warp to that template, and it can map accompanying datasets (from subj → template space, or vice versa) with that warp automatically. Finally, its outputs can be put into afni_proc.py for creating a full FMRI processing pipeline.
–pt
wow, this looks pretty good  . Thank you for this. I am trying to repeat the results as you suggested, but have not succeeded so far. So, if you can share the code you used, I would appreciate it.
. Thank you for this. I am trying to repeat the results as you suggested, but have not succeeded so far. So, if you can share the code you used, I would appreciate it.
Yes, I will send you the dataset I used as a reference template. At the moment, this is a “work in progress” on fixing the properties of this template and its accompanying atlases, masks, etc. But this first draft of sorting out the template’s properties seems OK.
The command I used (in tcsh scripting syntax) was as follows (the $run variable isn’t so important, but I wasn’t sure if I would have to test different options, and I wanted to keep track of each output and log file separately—as it happened, just the defaults worked fine):
#!/bin/tcsh
set subj = SUBJ_ID
set anat = DSET_NAME
set run = 00
set template = canfam_v3_template.nii.gz
@animal_warper \
-input ${anat} \
-input_abbrev ${subj}_anat \
-base ${template} \
-base_abbrev CANFAM3 \
-outdir out_aw_${subj}_${run} \
|& tee log_aw_${subj}_${run}
–pt
I have succeeded to strip the skull and found the reason for the previous failure. I need to use the ‘3dresample -master ${template} …’ to resample the original anat to the grid of the template first and then to run the @animal_warper program. Thanks again !
Hi-
I am glad things are working for you, but I do not see why you would need to resample the the original anatomical image to the grid of the template. That should not be necessary, and it would only (likely) smooth your input anatomical.
I don’t understand, because I did not resample the anatomical you had given me, and @animal_warper worked fine.
Can I ask what error you were getting before you used 3dresample?
–pt
I am pretty confused about our results looking different.
Can I ask what version of AFNI you are using—what is the output of “afni -ver”?
–pt
I’m also confused about this. I’m using AFNI_21.2.01
I don’t see any changes to @animal_warper since that time.
I have processed the dataset you sent me on both Ubuntu and Mac OSs, and the results come out the same (good) each time, without resampling. What OS are you using? And can you please copy+paste here the exact command you are running, again, just to be sure?
And finally, would you mind please updating your AFNI:
@update.afni.binaries -d
… and see if that makes a difference?
Thanks,
pt
Hi-
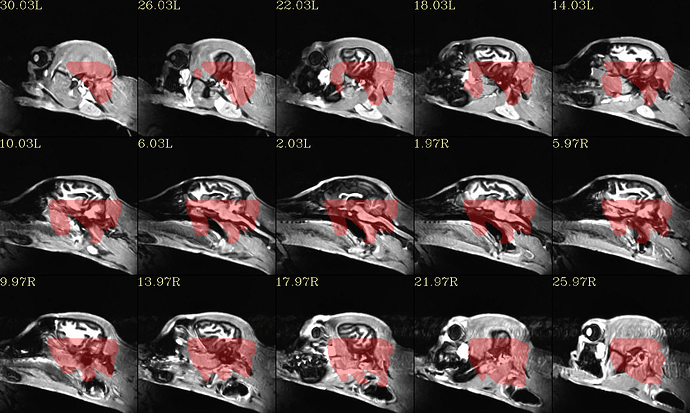
Another thing I notice, we have each uploaded the equivalent QC image made by @animal_warper (qc_02*sag.png), but the FOV looks different between them—you can see the FOV includes a loooot of space around the brain in the QC image I uploaded, but in the one you uploaded, it is much tighter around the brain. So, somehow I think something is different with the input dataset being used. I used the one in the “anat/” directory in what you shared with me.
The reason this might affect things is that if the starting coordinates are very different, the initial alignment may be off and some option to handle this might be required. That might be why resampling to the template helped your output—but there are probably other ways to fix bad coordinates that won’t risk smoothing the data.
Please let me know what dataset you are using, perhaps sharing it if it is a different one than what I had had.
–pt
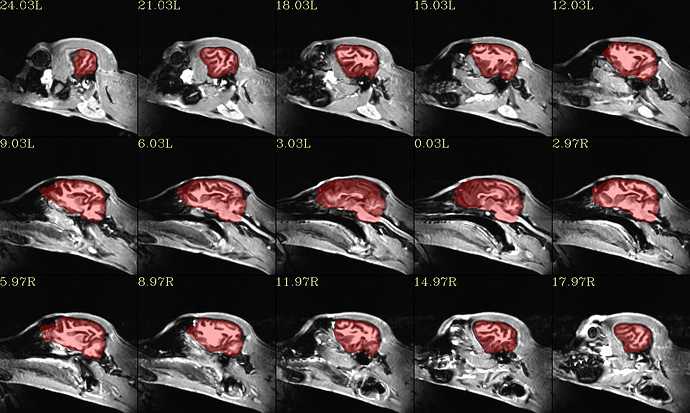
Avoiding to be confused, I rerun the following program using the anatomical from Google Drive I shared with you.
#!/bin/tcsh
set subj = Hali
set anat = /mnt/j/huotengbin/Dog_project/20210710_Hali_RS_husky1-4/T1Img/Hali/T1_den_N4_from_GoogleDrive.nii
set run = 00
set template = /mnt/j/huotengbin/Dog_project/Johnson_etal_2019_Canine_Atlas_v2/Canine_population_template.nii.gz
@animal_warper \
-input ${anat} \
-input_abbrev ${subj}_anat \
-base ${template} \
-base_abbrev Johnson \
-outdir out_aw_Johnson0823_noresample_GD_${subj}_${run} \
|& tee log_aw_Johnson0823_noresample_GD_${subj}_${run}
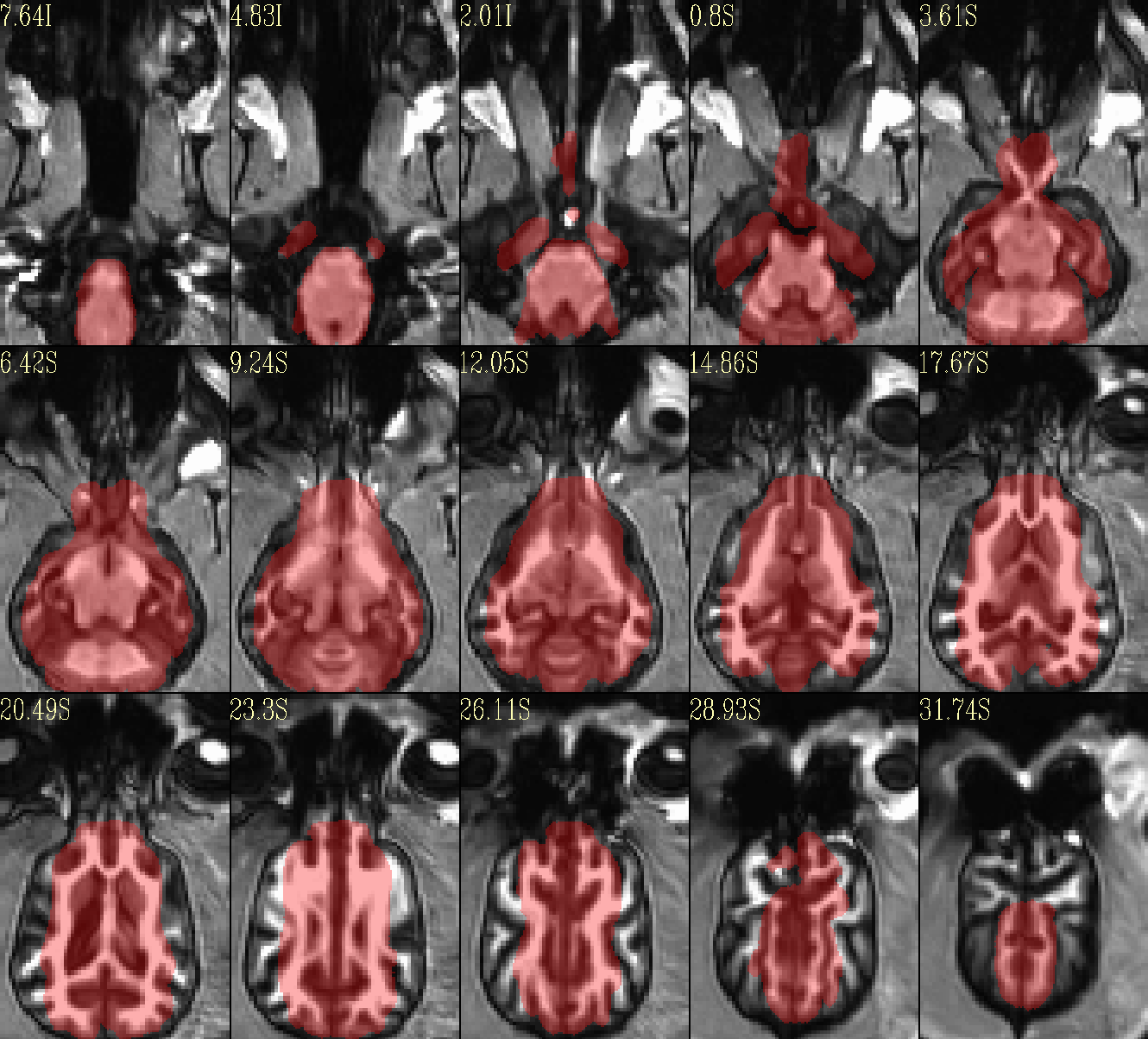
I got the following result.
Then, I aslo applied resampling to the see the difference.
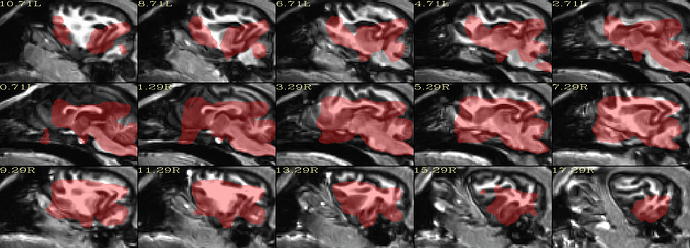
#!/bin/tcsh
set subj = Hali
set anat = /mnt/j/huotengbin/Dog_project/20210710_Hali_RS_husky1-4/T1Img/Hali/T1_den_N4_from_GoogleDrive.nii
set anat_resample = /mnt/j/huotengbin/Dog_project/20210710_Hali_RS_husky1-4/T1Img/Hali/T1_den_N4_from_GoogleDrive_res.nii
set run = 00
set template = /mnt/j/huotengbin/Dog_project/Johnson_etal_2019_Canine_Atlas_v2/Canine_population_template.nii.gz
3dresample -master ${template} -prefix ${anat_resample} -input ${anat}
@animal_warper \
-input ${anat_resample} \
-input_abbrev ${subj}_anat \
-base ${template} \
-base_abbrev Johnson \
-outdir out_aw_Johnson0823_resample_GD_${subj}_${run} \
|& tee log_aw_Johnson0823_resample_GD_${subj}_${run}
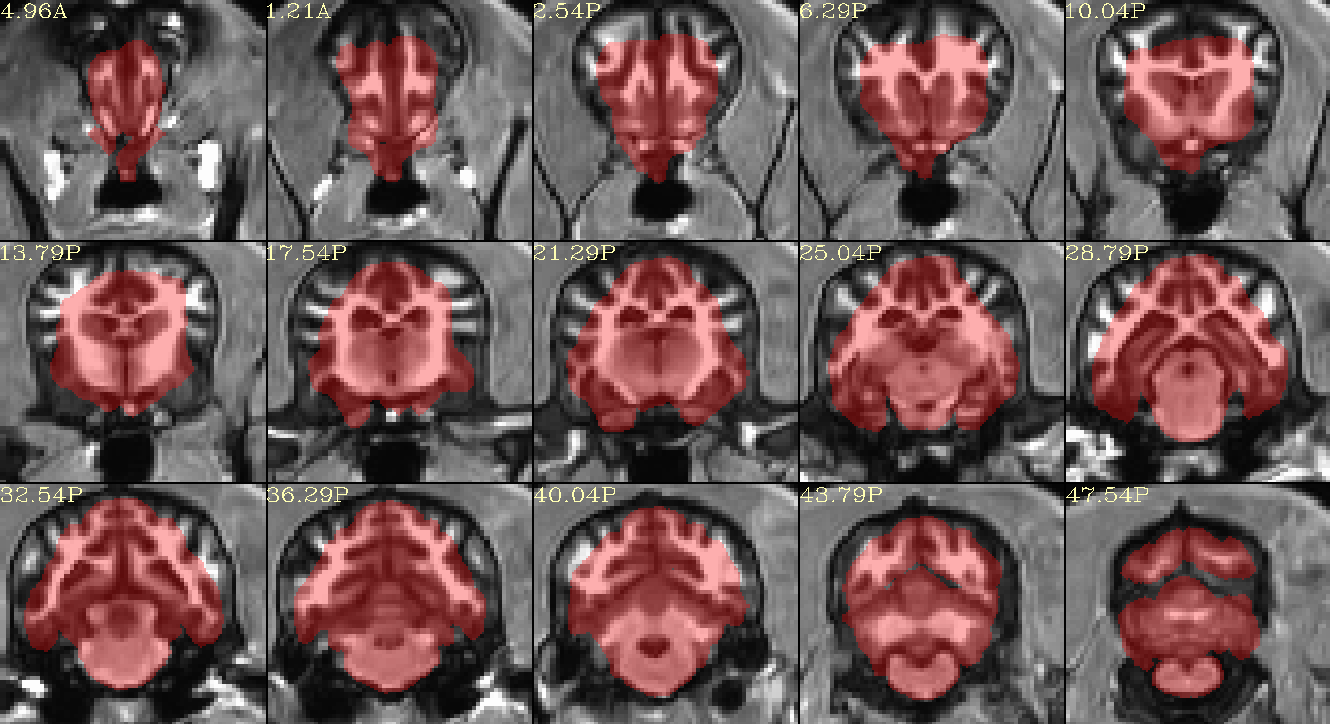
The results like:
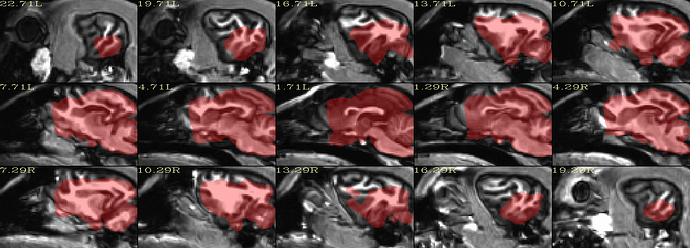
When using another template (A high-resolution MRI brain template for adult Beagle - PubMed), I can get a better result. I have uploaded the template to Google Drive, you can download for use.
#!/bin/tcsh
set subj = Hali
set anat = /mnt/j/huotengbin/Dog_project/20210710_Hali_RS_husky1-4/T1Img/Hali/T1_den_N4.nii
set anat_resample = /mnt/j/huotengbin/Dog_project/20210710_Hali_RS_husky1-4/AFNI_processed_v5/T1_lxr.nii
set run = 00
set template = /mnt/j/huotengbin/Dog_project/M30template/T1_Template_M30mask.nii
set template_resample = /mnt/j/huotengbin/Dog_project/M30template/T1_Template_M30mask_lxr.nii
3dresample -dxyz 1 1 1 -prefix ${template_resample} -input ${template}
3dresample -master ${template_resample} -prefix ${anat_resample} -input ${anat}
@animal_warper \
-input ${anat_resample} \
-input_abbrev ${subj}_anat \
-base ${template_resample} \
-base_abbrev lxr_mask \
-outdir out_aw_lxr_${subj}_${run} \
|& tee log_aw_lxr_${subj}_${run}