Dear Neurostars Members,
I have processed our multi-echo resting state fMRI data with fMRIPrep and tedana and would like to investigate the potential tSNR enhancement of the optimally combined image versus the second echo of the aquired images.
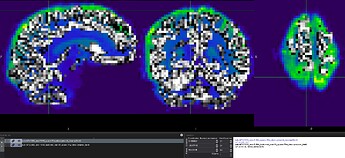
Images are processed initially with fMRIPrep and fed to tedana for further processing and ICA denoising. The optimally combined image I used for the tSNR computation is sub-MTL0002_ses-FU84_task-rest_run-01_space-T1w_desc-preproc_bold.nii.gz and sub-MTL0002_ses-FU84_task-rest_run-01_echo-02_desc-preproc_bold.nii.gz as the preprocessed second echo image generated by fmriprep. Based on this fomer topic on neurostarts, I generated the tSNR map of these images.
Based on this paper and in line with several others, I assume the following appraoch would be a reasonable one to find a measure of tSNR for comparison: “Quantifying tSNR within the conjunction of grey matter and functional masks. Resampling skull-stripped anatomical images to functional resolution and segmenting to create the grey matter mask. Using the median of all voxels within this mask”.
Accordingly, I resampled sub-MTL0002_ses-FU84_acq-MPRAGE_label-GM_probseg.nii.gz to the functional image (sub-MTL0002_ses-FU84_task-rest_run-01_space-T1w_desc-preproc_bold.nii.gz) using AFNI with the bellow command:
Resampling command:
3dresample \
-master sub-MTL0002_ses-FU84_task-rest_run-01_space-T1w_desc-preproc_bold.nii.gz'[0]' \
-inset sub-MTL0002_ses-FU84_acq-MPRAGE_label-GM_probseg.nii.gz \
-rmode Linear \
-prefix sub-MTL0002_ses-FU84_space-T1w_desc-gmprob_resamp2bold.nii.gz
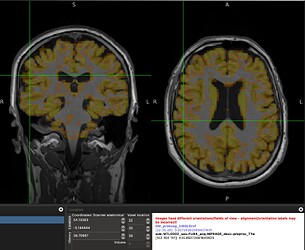
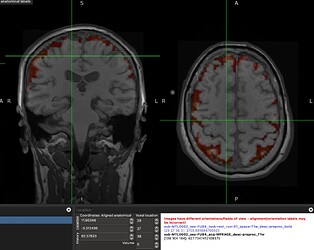
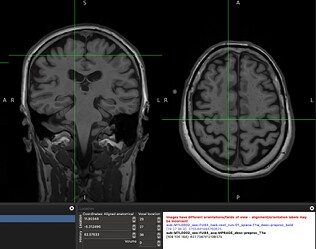
When overlapping the resampled GM mask on the functional image, they do not perfectly overlap and the mask misses the high signal regions. This conserns me of how accurate would be the median of tSNR values laying within this mask?
A screenshot of the misalignment:
I’d appreciate any guidance on resolving this issue or having an alternative approach to compute the tSNR maps within the GM for the above purpose.
Many thanks,
Ali