Hey,
I found some very strange results and came here in the hope that someone might be able to explain it. I classified which task a subject was executing in a task-switching paradigm using whole-brain fMRI data. I used sklearn.svm.SCV for classification as well as nilearn.decoding.SpaceNetClassifier. Both yielded good performance, but as expected very different weight vectors.
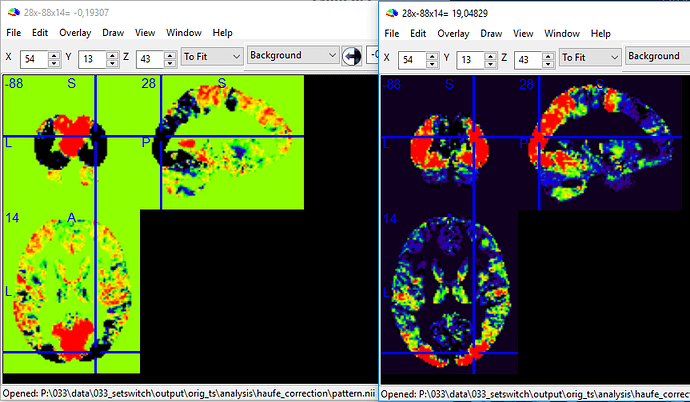
Then, I corrected the weigth-vectors according to Haufe et al. (2014) using the posthoc library to make interpreting them a valid approach. Which, for this scenario, boils down to multiplying the weight-vector with the covariance matrix of the features; if I understood it correctly. The fascinating result was, that after the correction both images looked very similar (which is awesome), but were inverted:
As the input for both classifiers was identical for both, features and labels (which were 0 and 1) this is very unexpected. I looked at the code for both classifiers. The SVM preserves the labels while the SpaceNet maps the 0 to a -1. However, I don’t see how that explains the inverted results.