Hello,
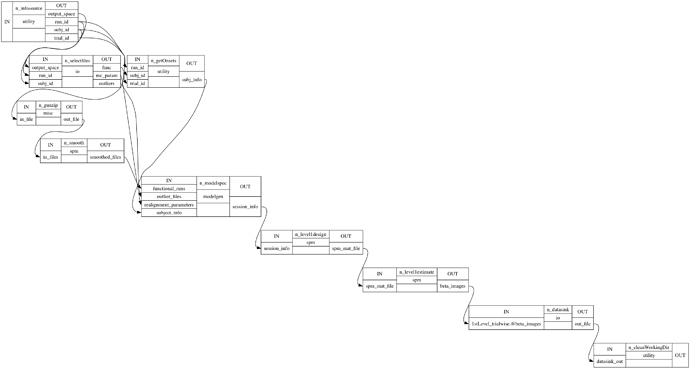
I need trial-wise beta estimations for my analysis. For this I have setup runs and trials as iterables. So for each subject, run and trial I compute a GLM and save the beta images. This creates a huge amount of data in the working directory because each node saves the data (e.g. functional .nii files for each run) in a separate folder.
I know there is the experimental config option to delete node folders, but this crashes the workflow as SPM is looking for a file that is not there (my guess is that it gets deleted but is actually still needed).
As a workaround I have tried to connect a self-written function to the output of the datasink. This way I know that the workflow for the specific subject, run and trial has finished. In my function I then delete the node folders for that specific subject, run and trial. However, the workflow still has errors because nodes try to access files that are already deleted.
I came here to ask if anybody else has tried to clean up the working directory “on the fly” and might be able to give me some advice. Thanks!