Summary of what happened:
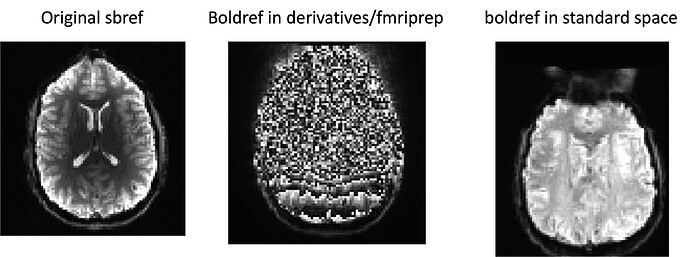
We ran fmriprep successfully, on a dataset with a T1, task data, and single band reference images. The subject’s sbrefs look fine in the initial form (in bids/[subj]/func/), but the intermediate image in bids/derivatives/fmriprep/[subj]/func (named with suffix boldref) is essentially all noise. Then the boldref image in standard MNI space looks OK again (see attached image).
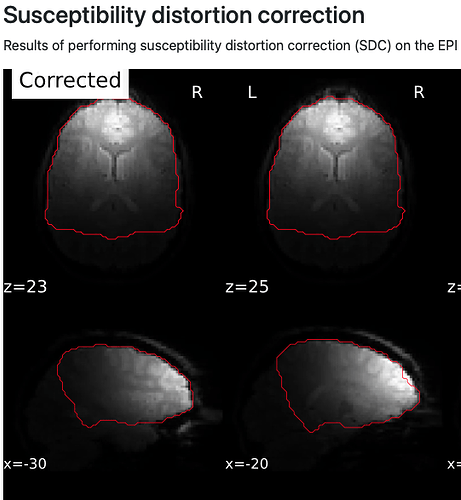
Relatedly (I assume), the brain mask shown in the susceptibility distortion correction part of the QC report is messed up, missing the posterior third of the brain (see attached image).
I’m trying to trace back where the problem originates, and am having trouble finding all the commands, and the order in which they’re run. Is there a place where fmriprep stores a log of the actual commands, and not just the workflows? I see “command.txt” files in some, but not all the *_wf folders. If the command.txt file doesn’t exist, does that mean no commands were run, or is there another place where the command is saved? I would just like to reproduce the problem in as minimal a way as possible (not run fmriprep again).
Aside from that, does anyone know what might cause this problem and how to fix it? Other runs from the same session were fine, so maybe it’s just motion, but I know it’s not something inherent to the scan session.
From digging around it seems that the initial affine transform (calculated by antsAI) in func_preproc_task_[taskname]_wf/final_boldref_wf/enhance_and_skullstrip_bold_wf/init_aff might be the culprit, but I’m also seeing this comment in the fmriprep documentation on the bold_reg_wf:
"
After either BBR workflow is run, the resulting affine transform will be compared to the initial transform found by FLIRT. Excessive deviation will result in rejecting the BBR refinement and accepting the original, affine registration.
"
I thought this may explain why the final boldref image in standard space is ok (because it rejected the bad affine transform?) but I couldn’t find where to check whether the BBR refinement was accepted or rejected.
Command used:
docker run --rm -it -v bulk:bulk \
-v scratch:scratch \
-v /usr/global/freesurfer:/usr/global/freesurfer \
nipreps/fmriprep:23.0.2 \
bulk/study/bids \
bulk/study/bids/derivatives/fmriprep \
participant \
-w scratch/study_fmriprep/subj1 \
--participant-label subj1 \
--nprocs 8 \
--random-seed 2312 \
--skull-strip-t1w force \
--force-bbr \
--slice-time-ref 0 \
--fs-subjects-dir bulk/study/bids/derivatives/freesurfer \
--fs-license-file /usr/global/freesurfer/6.0.0/license.txt \
--write-graph --verbose
Version & Environment:
Running fmriprep 23.0.2 on Docker