Summary of what happened:
I used qsirecon 1.1.2.dev0+gfa8673957.d20250819 and the mrtrix_multishell_msmt_ACT-fast workflow to compute adjacency matrices for my subjects. Everything worked fine, but now I need to compute a group average adjacency matrix. For this it is necessary to normalize the matrices in such a way that I get rid of absolute differences between subjects (e.g. in term of total fiber counts, etc).
For the connectivity measure, I decided to stick with sift_radius2_count_connectivity for now, as both the radius and sift corrections seem reasonable to me (However, I am not bound by this metric, I could also choose another one if this helps with my issue)
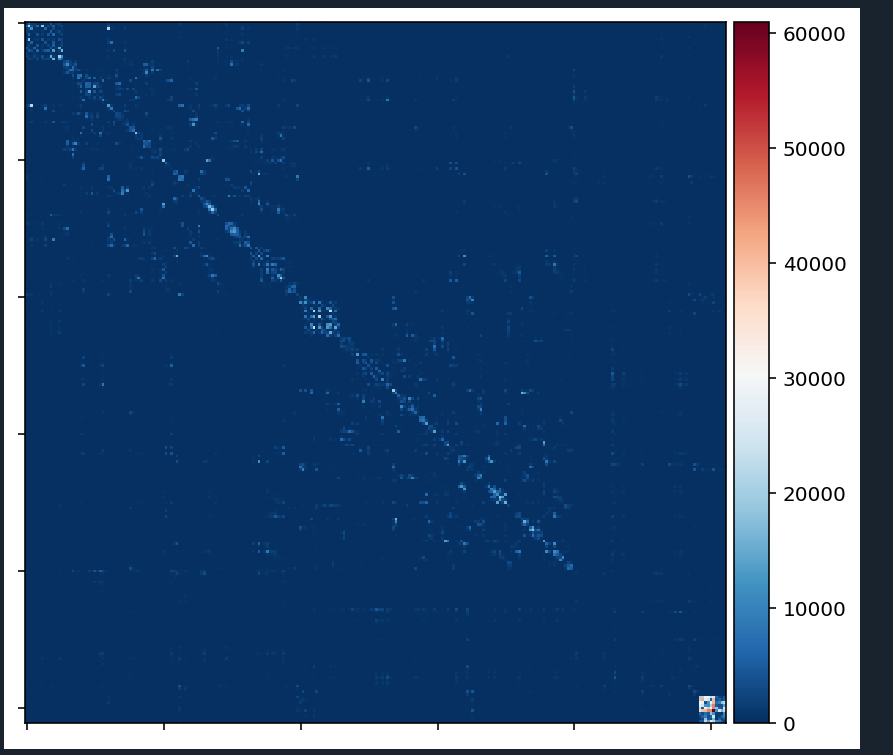
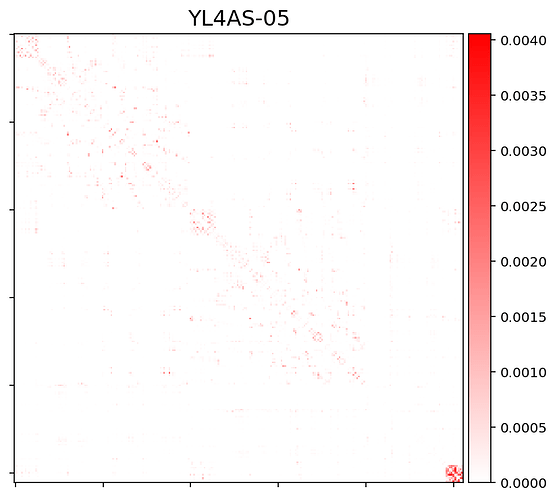
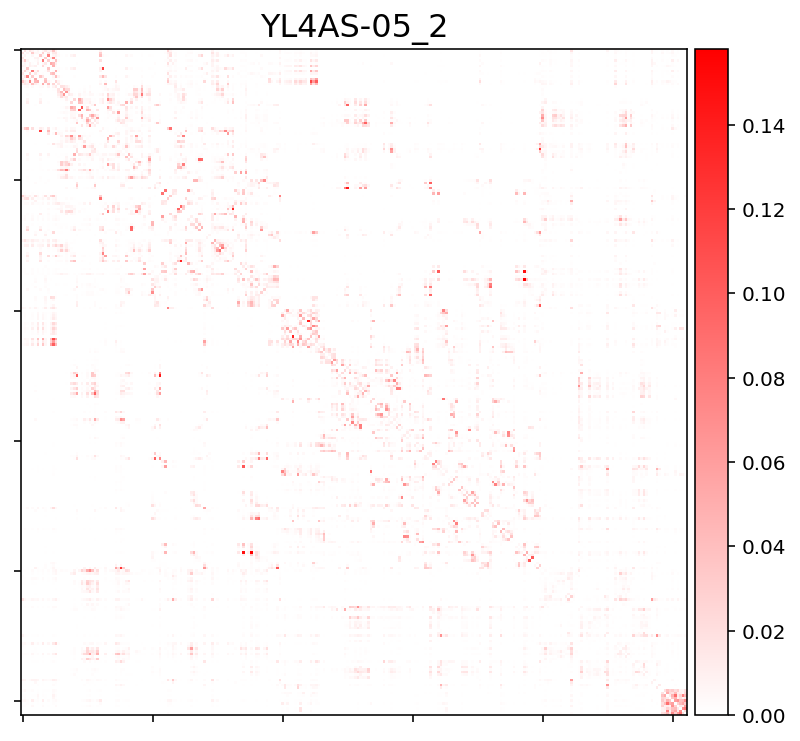
For now I only set the diagonal to 0 (to exclude self-connections, which is needed for the downstream analysis). Then I plotted the matrices using nilearn.plotting.plot_matrix.
Problem: Currently it seems that the matrices are heavily biased towards within-subcortical and/or within-cerebellar connectivity.
I wonder:
1.) If and how I could process the matrices in such a way that they are more “balanced” (within subject)
2.) How should I then should compute the group-average matrix (might or might not be related to 1., so can be ignored if 2. does not depend on 1.)
This might be related to this thread?
Command used (and if a helper script was used, a link to the helper script or the command generated):
I only set --recon-spec mrtrix_multishell_msmt_ACT-fast (otherwise just defaults)
Version:
qsirecon 1.1.2.dev0+gfa8673957.d20250819
Screenshots / relevant information:
This is how the connectivity matrix for one subject currently looks like. Note:
It does not matter which connectivity measure I take (the matrix will more or less look the same). So I get a similarily looking matrix with:
sift_radius2_count_connectivity
radius2_count_connectivity
sift_invnodevol_radius2_count_connectivity
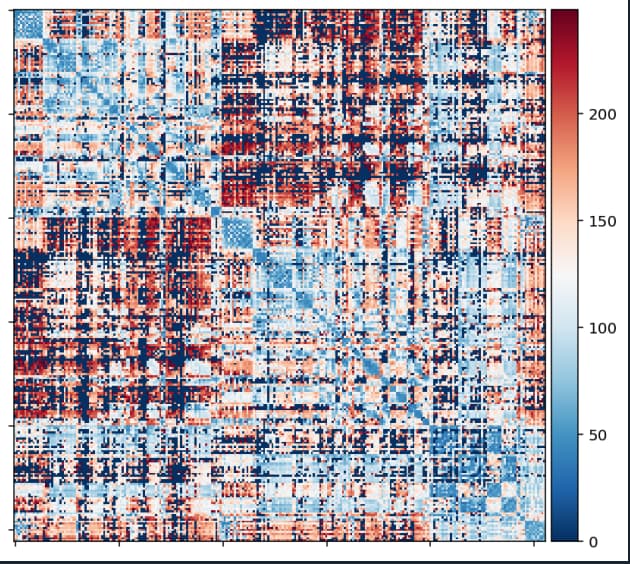
However, the matrix for radius2_meanlength_connectivity looks much “better”: