Is it possible to add the older adult MNI MRI template discussed here https://www.ncbi.nlm.nih.gov/pubmed/22440645 to fmriprep? Nii format file comes with the following clinical toolbox: https://www.nitrc.org/projects/clinicaltbx/
Thank you,
Jeremy
As one of the authors behind the clinical toolbox, I would be delighted if people used my work and cited my paper. However, users should be aware that the template developed by Rorden et al. (2012) was specifically designed for use with SPM. While generally ignored, it should be noted that SPM tends to generate average sized brains, while tools like FSL and ANTs come with templates that are MNI template sized. The MNI template is larger than average. As noted when the template was released, this was a side-effect of the algorithm used to develop this template (there is no evidence that people in Montreal have larger than average brains). This difference is best seen in Figure 1 of Horn et al. (2017). This difference explains why MRIcroGL’s File/OpenStandard menu lists mni152 as well as spm152. The former template is for images normalized to MNI sized brains, while the latter is for average sized brains.
As you note, human brain volume shrinks on average about 5% for each decade after 40. Therefore, it might be worthwhile using an older template.
I really do not know how the regularization tuning influences different tools. I do know the images developed by Rorden et al were intentionally designed to work hand in glove with SPM. For more general algorithms, I suspect there are better options, e.g. those from John E. Richards
This is great input. Thanks so much Chris! Given this, it does seem better to have these older adult templates added as an option in fmriprep instead.
Hi @Jeremy_Purcell, I have added https://github.com/templateflow/templateflow/issues/19 with your request.
@Chris_Rorden, do you happen to remember the license of the template and whether it would allow us to redistribute it (of course requesting the citation of whatever works need be referenced).
I have a set of average MRI templates. Range in age from 2 weeks through 89 years, in 3 mo, 6 mo, half year, or five year intervals. I would be glad to have someone add them to the fmriprep processing. The avg templates are hosted on NITRC under neurodevelopmental mri database, group is neurodev. Chris referenced my www site that has info. John
Thanks, do you know when this template will be added? I am interested in using it for my current data analysis…
Hi there!
I am also very much interested in using an older adult template for my analysis. I am actually investigating a study cohort made up of older adults, and individuals with mild cognitive impairments and Alzheimer’s disease, and so a template made from this cohort would be even better, if possible.
I just wanted to double-check that the only way in order to use custom templates which are not yet available, is that we would need to upload them to tensorflow and allow them for shared public usage? Currently, is there is no function within fmriprep which allows users to use templates that are stored locally/privately? With our study protocols, I am unsure whether our data can be uploaded and shared publicly due to our ethics.
You can your custom templates without sharing (or uploading anywhere). Just add your template to the TemplateFlow (not tensorflow) home directory. Check Python Client - TemplateFlow docs.
Once your template is indexed by TemplateFlow, you just pass your template name with the --output-spaces argument (e.g., --output-spaces MyCustomTemplateName).
A custom template is not identifiable data, so very unlikely sharing it would be that cumbersome. But yes, you should definitely see whether you can publish derived data under a local ethics approval.
These templates are not publicly available, so the only way for you is to proceed to the NITRC page and fill in all the required information, get approved by the owner of the team, download the data, re-structure it following TemplateFlow’s guidelines and then add it as a custom template as I mentioned above.
Alternatively, someone could upload Chris’ template (acknowledging the problems he mentioned in his post) to TemplateFlow. Unfortunately, I don’t have the bandwidth to do this right now.
Thanks so much!
I installed templateflow and changed the path of the environment variable, $TEMPLATEFLOW_HOME. I have added my custom template into the templateflow directory and specified the output space as --output-spaces DPRCcustom. However upon running fmriprep, I get an error saying that the ‘tpl prefix’ is required, but have made sure to include the tpl- prefix on the folder and file name. My custom template directory looks like this:
$TEMPLATEFLOW_HOME/
tpl-DPRCcustom/
template_description.json
tpl-DPRCcustom_T1w.nii.gz
I am unsure of what the problem is, and would appreciate any input, thanks!
- How are you running fMRIPrep? (or more precisely, are you using containers? and what is the command line?
- I believe fMRIPrep requires your template to have a corresponding brain mask (probabilistic or binary, I think the former has more priority).
Hi,
Any update on this matter?
I’ve created my own custom template with related brain mask (I used names convention as in the others templatefow folder - MNI template like style). Then, I copied them into the TEMPLATEFLOW_HOME (using prefix tpl- for both folder and filenames).
Filenames in the template flow folder (tpl-Custom23Subj) are:
template_description.json tpl-Custom23Subj-brain_mask.nii.gz tpl-Custom23Subj_T1w.nii.gz
However, I got the error: –skull-strip-template: invalid from_string value: ‘tpl-Custom23Subj’.
This is the command I’m using (fmriprep-docker version is 20.10.16):
fmriprep-docker $bids_root_dir $outdir participant --participant-label G1001
–bids-database-dir $outdir
–task rest
–nthreads $nthreads
–omp-nthreads 4
–mem_mb $mem_mb
–md-only-boilerplate
–fs-license-file $FS_LICENSE
–fs-no-reconall
–ignore flair
–skull-strip-template Custom23Subj
–output-spaces MNI152NLin6Asym:res-2:res-1 MNI152NLin2009cAsym tpl-Custom23Subj
–force-bbr
–topup-max-vols 1
–use-aroma
–error-on-aroma-warnings
–stop-on-first-crash
–verbose
–work-dir $workingdir
Is this related to the fact that fmriprep is not seeing properly the templateflow folder?
Other notes that might be important:
I have enabled datalad (as reported here Python Client - TemplateFlow).
If I run on already available standard template (MNI152NLin6Asym and MNI152NLin2009cAsym) it is working without errors.
Thank you in advance for any help.
Best,
Riccardo
Hi, you will need to mount the templateflow folder where you wrote your custom template into the container.
Hi,
Thank you for your reply.
I did several attempts with both the fmri-docker wrapper and by invoking directly the docker command. I prefer the latter where it is easier to understand how to mount directories.
Here it is the command:
docker run --rm -e DOCKER_VERSION_8395080871=20.10.16 -it
-v /usr/local/freesurfer/license.txt:/opt/freesurfer/license.txt:ro
-v $bids_root_dir:/data:ro
-v $outdir:/out
-v $workingdir:/scratch
-v $TEMPLATEFLOW_HOME/tpl-Custom23Subj:/home/fmriprep/.cache/templateflow/tpl-Custom23Subj:ro
nipreps/fmriprep:21.0.2 /data /out participant --participant-label G1001
–task rest
–nthreads 8
–omp-nthreads 4
–mem_mb 50000
–md-only-boilerplate
–fs-no-reconall --ignore flair
–topup-max-vols 1 --error-on-aroma-warnings
–stop-on-first-crash --use-aroma --verbose --sloppy
-w /scratch --output-spaces MNI152NLin2009cAsym Custom23Subj:res-01
However, this is the error:
FileNotFoundError: No such file or directory ‘[]’ for output ‘t1w_file’ of a TemplateFlowSelect interface
Probably, is the mounting to the tempalteflow dir not working correctly? I do not have ideas on how to fix it.
Thank you.
Riccardo
To confirm whether the folder is there:
docker run --rm -it \
-v /usr/local/freesurfer/license.txt:/opt/freesurfer/license.txt:ro \
-v $bids_root_dir:/data:ro \
-v $outdir:/out \
-v $workingdir:/scratch \
-v /path/to/hosts/folder/tpl-Custom23Subj:/home/fmriprep/.cache/templateflow/tpl-Custom23Subj:ro \
--entrypoint=/bin/bash \
nipreps/fmriprep:21.0.2
and then, at the prompt:
$ echo $TEMPLATEFLOW_HOME
$ ls -lah ${TEMPLATEFLOW_HOME:-/home/fmriprep/.cache/templateflow/}/*
Thank you again for your help.
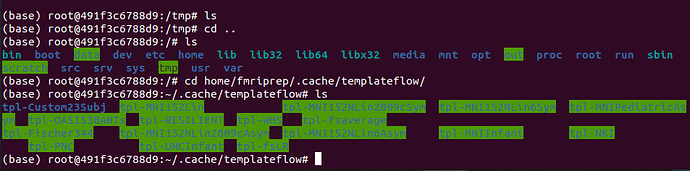
I did as you suggested and the folder seems to be properly mounted into the docker under the directory /home/fmriprep/.cache/templateflow.
Also, the command ls -lah is printing out all the template folders in templateflow, including my custom template.
However, I got the following errors:
traits.trait_errors.TraitError: The ‘t1w_file’ trait of a _TemplateFlowSelectOutputSpec instance must be a pathlike object or string representing an existing file, but a value of ‘[]’ <class ‘str’> was specified
and another exception:
FileNotFoundError: No such file or directory ‘[]’ for output ‘brain_mask’ of a TemplateFlowSelect interface
Thank you.
Riccardo
Can you copy and paste the output of the two command lines I suggested?
Then, what’s the output of:
docker run --rm -it \
-v /usr/local/freesurfer/license.txt:/opt/freesurfer/license.txt:ro \
-v $bids_root_dir:/data:ro \
-v $outdir:/out \
-v $workingdir:/scratch \
-v /path/to/hosts/folder/tpl-Custom23Subj:/home/fmriprep/.cache/templateflow/tpl-Custom23Subj:ro \
--entrypoint=/opt/conda/bin/python \
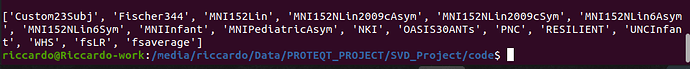
nipreps/fmriprep:21.0.2 -c "from templateflow import api; print(api.templates())"
Sure. You can find them attached.
first_command.png contains a screenshot where I navigated into the templateflow folder mounted into the docker (it opened in the #tmp folder which is empty). It seems that the tpl-Custom23Subj folder is there.
second_command.txt contains the output of the echo and ls -lah command.
second_command.txt (158.2 KB)
third_command.png is the new one you suggested in the last post.
Finally, I also attached the output of the full fmriprep command with the errors. Maybe it can help.
fmriprep_command.txt (18.1 KB)
Thank you.
Riccardo
Hi,
I know you are very busy, but I hope you can still help me solving this issue.
Thank you,
Riccardo
I am getting the same error as @Ric when trying to use a custom template:
220916-15:24:38,890 nipype.workflow WARNING:
[Node] Error on "fmriprep_wf.single_subject_SA27_wf.anat_preproc_wf.anat_reports_wf.tf_select" (/ptmp/aenge/tmp/work_job_5924972/fmriprep_wf/single_subject_SA27_wf/anat_preproc_wf/anat_reports_wf/_template_MyTemplate/tf_select)
220916-15:24:38,894 nipype.workflow ERROR:
Node tf_select.a1 failed to run on host co2643.
220916-15:24:38,894 nipype.workflow ERROR:
Saving crash info to /ptmp/aenge/tmp/ds_job_5924972/derivatives/fmriprep/sub-SA27/log/20220916-152241_bd4f05af-c923-4fc9-9695-eae117507cba/crash-20220916-152438-aenge-tf_select.a1-f4256be3-9732-4ad7-b575-15c595850968.txt
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/traits_extension.py", line 129, in validate
value = Path(value) # Use pathlib's validation
File "/opt/conda/lib/python3.8/pathlib.py", line 1042, in __new__
self = cls._from_parts(args, init=False)
File "/opt/conda/lib/python3.8/pathlib.py", line 683, in _from_parts
drv, root, parts = self._parse_args(args)
File "/opt/conda/lib/python3.8/pathlib.py", line 667, in _parse_args
a = os.fspath(a)
TypeError: expected str, bytes or os.PathLike object, not list
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 530, in aggregate_outputs
setattr(outputs, key, val)
File "/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/traits_extension.py", line 131, in validate
self.error(objekt, name, str(value))
File "/opt/conda/lib/python3.8/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: The 't1w_file' trait of a _TemplateFlowSelectOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '[]' <class 'str'> was specified.
As for @Ric, the name of my custom template gets shown correctly in the output api.templates() and seems to have all the necessary files, as far as I can tell (tpl-MyTemplate_T1w.nii.gz, tpl-MyTemplate_desc-brain_mask.nii.gz, and template_description.json).
Any tips would be very much appreciated!