Hi, it looks like you are in the case of phase difference map, which is not a case where FSL topup is used but rather FSL PRELUDE as it can been seen here (look for function: init_phdiff_wf).
I don’t think that it is a problem to have your phase difference image with different phase encoding direction than in your functional images (usually phase difference images map are generated using a GRE sequence with no distorsion along the phase encoding direction).
For the phase different map correction implemented by fmriprep (details of the method are here) you may need to try to change “i” into “-i” in fmri json file to see if the correction is better.
How are your native functional images? Are the distorsions really along the L-R direction (meaning that the phase encoding direction is really “i” or “-i” )?
Which version of fmriprep are you using?
My understanding is that one set of images was acquired with AP axis and the other LR. The assumption of TOPUP and similar tools is that the truth lies precisely in the middle of the two phase reversed images (e.g. A>P and P>A or alternatively L>R and R>L). This axiom does not work if the image pair do not have 180 degree phase difference relative to each other (e.g. A>P and R>L). dcm2niix is faithfully labelling the directions as they were acquired. The images are not suitable for undistortion.
Thanks for this detailed report. Is there any chance you’d be willing to share the data? I’ve opened Potential issue when (phase difference) fieldmap PE orientation does not match EPI · Issue #325 · nipreps/sdcflows · GitHub where this issue can be tracked.
One diagnostic for whether this is potentially fixable is trying to run the latest fMRIPrep LTS release (20.2.7). That used a different architecture for susceptibility distortion correction, and will let us know if this is a regression. If that works correctly, there is a high likelihood we can track down the deviation and fix it in a later release. It is also possible that you have an unusable fieldmap, as Chris suggests, so I can’t make promises.
We do have a fieldmap-less SDC approach (use --ignore fieldmaps --use-syn-sdc) in the meantime, which will make a best effort to correct distortions in the phase-encoding direction, using the anatomical image as a reference.
Hi @Chris_Rorden, indeed, according the the initial post, the GRE-EPI sequence used for functional images seems to have been acquired in LR whereas the set of images acquired with the GRE recalled sequence seems to have been acquired with the phase direction along the AP direction. I use intensively topup for SDC of my EPI images (both for functional and diffusion) and I was thinking that the SDC with the phase difference map method (which is not based on the acquisition of two SE-EPI images with reverse polarity, but rather two GRE images with two different echo times) would work for a different phase encoding direction direction of the fieldmap with respect to the phase encoding direction of the functional scans.
As stated by @effigies, this may not be the case and that could be the source of the problem observed by @mkhazova . Thank you @effigies for starting this new issue, I will be on the lookout for this interesting topic.
As a side note, I read this part from the topup User guide, meaning that you don’t necessarily need to have your set of EPI images with exactly 180 degree phase difference relative to each other to work with topup, even if the latter is preferred in the general case:
Topup gives you lots of freedom in how to acquire your data and still be able to calculate and correct for distortions. You can e.g. use
1 0 0 0.087
0 1 0 0.087
indicating that the first scan was acquired with phase-encoding in the left-right direction and the second in the anterior-posterior direction. One can even acquire ones data like
0 1 0 0.087
0 1 0 0.107
i.e. with distortions going in the same direction in both acquisitions but with slightly larger distortions in one case (the latter in this example).
Despite this flexibility it is nevertheless best to manipulate the phase-encoding so that it yields the maximum difference in the observed images, and that will a reversal of the blips. So unless one has a very good reason to do otherwise that is the recommended type of acquisition.
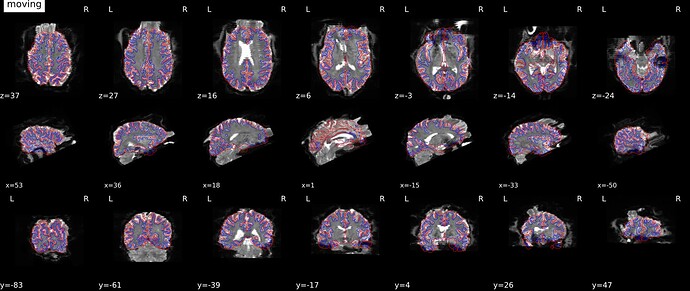
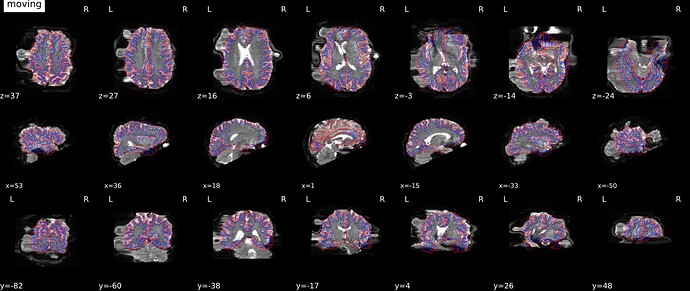
I’m very confident this doesn’t have anything to do with the phase encode direction, and is just a manifestation of the SDC regression that has been noted elsewhere. I am using the same technique: phase difference field map GRE acquisition with phase encoding i, and my bold acquisition has phase encoding j-. I have identical looking distortions with version 22.1.1 and 22.0.2, but the suggestion to try the LTS release (20.2.7) gave a good result. I posted screenshots showing a comparison of both versions on the same data here:
fmriprep-sdc-distort-the-image-and-there-is-error-for-some-subjects
I also tried manually changing the phase encode direction on the phase difference and magnitude fmap images from i to j-, this predictably had absolutely no effect (ran on version 22.1.1, still looked really bad).
For me this artifact is super consistent, I ran around 60 datasets and I would say about half had very severe artifacts. I’ll update when all 60 are done running on 20.2.7 but so far they look good.
I’m happy to help by sharing more examples or data if you need it.
All 60 datasets have been completed with 20.2.7 with very good results. Again I can send example data of cases where the SDC regressed if that is helpful.
Yes, example data will be very useful. However is best for you to share it, you can send a link to this username @ gmail.
Hello, dear colleagues.
I apologize for the long answer - I had to set up a virtual machine for data processing this files. I am new to working with the fmriprep, so I will write a sequence of actions. I use fMRIPrep 22.1.1.
Before running the script, I add the following lines to the json-files:
1. func:
“TaskName”: “buttonpress”,
“B0FieldSource”: “phasediff_fmap0”
2. fmap
- magnitude1:
“IntendedFor”: “func/sub-03_task-buttonpress_acq-TR1000_bold.nii”
- magnitude2:
“IntendedFor”: “func/sub-03_task-buttonpress_acq-TR1000_bold.nii”
- phasediff:
“B0FieldIdentifier”: “phasediff_fmap0”,
“IntendedFor”: “func/sub-03_task-buttonpress_acq-TR1000_bold.nii”
Then I run the script:
sudo fmriprep-docker /mnt/geesefs-mountpoint/PREPROCESS /mnt/geesefs-mountpoint//PREPROCESS/derivatives participant --participant-label 03 --fs-license-file /mnt/geesefs-mountpoint/freesurfer.txt
In order to fix the distortion, I tried to manually change the “i” to “j-” in the func json-file. I think it got better, but still not very good.
If I leave “i” (I do not change all the lines added earlier in json) and run the script:
sudo fmriprep-docker /mnt/geesefs-mountpoint/PREPROCESS /mnt/geesefs-mountpoint//PREPROCESS/derivatives participant --participant-label 03 --fs-license-file /mnt/geesefs-mountpoint/freesurfer.txt --ignore fieldmaps --use-syn-sdc
Did I do everything right and is there still a chance to correct this data?
Many thanks, Maria
Hi,
For this kind of fieldmaps, users seem to have better luck with fmriprep LTS v20.2.7:
Here is an example of such report:
Hi, I know this topic was over a year ago, but I would like to ask the source of that Siemens bug, or an official report. I am currently facing the same problems, and I was previously informed about a potential bug in these scanners when I asked others about those discrepancies. Still, no one seems to have an official statement from the manufacturer. I saw this thread and thought of asking here. Thanks!
Hi @DianaTru , this bug in Siemens sequences is resolved (as far as I know) with Syngo software versions XA. Which Siemens software version do you have on your MRI machine?
Hey @jsein, thanks for your response. It is great that the bug is already fixed. I am not sure what software version we had. It was a Siemens skyra (if that is of help). The thing is, I am working with legacy data, meaning it was acquired between 2016 and 2019, apparently while the bug was there, and we had two phase encode directions (PA and RL) in a sample of 200+ subjects. My main concern is that I have not found a legitimate source indicating the bug (as I said, officially addressed by Siemens) or nothing about the fix. Can you point me out on sources? Also, do you know if the data can be preprocessed as a single group (especially fMRI) given the two directions?
The scanner model doesn’t say much but the dates of acquisition make it very likely that the scanner version must have been VE or VD that were affected by this bug.
I don’t know any official source to document this bug. The fix was to copy the sequence in AP and manually rotate the phase encoding direction into PA, not relying on the “copy parameter” capability of the software as mentioned previously by @Chris_Rorden . As some point before the fix was made, there was a check box “copy phase encoding direction” that I think was more robust toward this bug.
In your case, it would be interesting to know how the protocol was saved at the console, and how it was run. Do you mean that the bold runs have either PA or RL phase encoding directions? What kind of fieldmap was acquired for theses subjects?
Actually, I am wrong, it seems that this glitch is also reported in XA30.
See here: http://www.bmap.ucla.edu/docs/PrismaUserGuide.pdf
Prisma XA30 Software Bugs
This is a list of known software bugs/glitches. If you encounter any other suspected issues,
please report them to bmctechs@mednet.ucla.edu and during study reporting.
AP/PA Copy Reference Issue
If you are using Siemens product sequences with different phase encoding directions (e.g. AP
then PA), you MUST NOT use the copy reference option. This glitch will change your second
PA sequence to RL. This glitch does not affect CMRR, HCP or ABCD sequences as the phase
encoding direction is controlled on the special card instead of the routine parameter card.
Workaround:
- You must manually input the angle for each sequence if you are manually prescribing
your FOV - If you use autoalign and DO NOT manually adjust your FOV in any way, then your scans
will not be affected and there would be no need to use copy reference
***This glitch has been reported to Siemens, but there is no way to know when and if
there will be a fix.
Thanks for looking deeper into this. I am not sure yet about the software version. Looking in the json files I found a line SoftwareVersions": “syngo MR D13”. Also, the scanner is a Skyra, so it seems the bug, particularly causing the flip from PA to RL, was present in other models. I was informed that a field map was not acquired for this dataset (I am unsure about the reason for such a decision). Now, when performing dicom to nifti conversion using heudiconv, I got a fmap folder with a “dir-AP_epi” json file. But I think there is no real field map. For this particular dataset, my problem is that in some subjects MPRAGE and BOLD sequences have different phase encode directions (e.g., BOLD has “i” and MPRAGE has “j”). In your expertise, is this something that can happen?, and are can I compare (or combine as part of the same group) subjects that have just PA direction?
This means that the software version was VD13, the bug was present.
If there is no nifti image, this means that there is no data to use to calculate a fiedlmap and corrrect your bold images for susceptibility distorsion correction (SDC) .Otherwise, if your bold runs are with dir-PA and if there one volume with dir-AP in the map folder, then you could do SDC for such a dataset, with methods such as topup (FSL).
You don’t have to worry about the MPRAGE images as these are images acquired with very different sequences. MPRAGE sequence is a 3D acquisition whereas bold EPI images are 2D axial acquisition. If you MPRAGE has “j” , I would guess there the sequence must a been prescribed in coronal, meaning the the readout direction must have been in “IS” direction, the first phase encoding direction in “AP” and the second phase encoding direction in “RL”. But in general you don’t have to worry about the phase encoding direction of the MPRAGE images since usually you don’t do SDC for MPRAGE images.
In your case, if for your 200 subject you have a mix a PA and RL (which means the the Phase EncodingDirection" reads “i” (P>A) or “-j” (R>L) ) , I would advise to do SDC for this data with tools such as SynBOLD-DisCo. SDC is in general recommended for BOLD analyses. See http://linkinghub.elsevier.com/retrieve/pii/S1053811902912814this paper for example, even more for in your case where the images are distorted differently from subject to subject due to different acquisition settings with different phase encoding directions.
Ok, not good for the studies acquiring large dataset on those days, but thanks for confirming.
Thank you so much for this! This is probably where I needed to go, and I didn’t know exactly how to approach it. I am particularly concerned on how mixed encoding could introduce inconsistencies across subjects, affecting both data reliability and spatial fidelity. One last question: I am curious if there could be effects in the registration into structural data, identification of resting-state networks, and mapping of regions of interest given this variability, particularly since I have been told that it is better to approach the data as two separate groups (reducing sample size as a consequence). Do you know if the methods to apply targeted correction techniques, or harmonization strategies for distortion alignment (either across RL and PA groups) reduce the chances to affect statistical power in group-level analysis?
It is an interesting topic. One could look at the output metrics for all your subjects and see if the measures could be clustered in two groups, based on the phase encoding direction of the acquisitions. I am not an expert there, but if you think at this issue as an harmonization issue as it is done across sites, you may try statistical method such as Combat to harmonize your data.