Hi,
thank you for all the advice. With the suggested adaption it worked now.
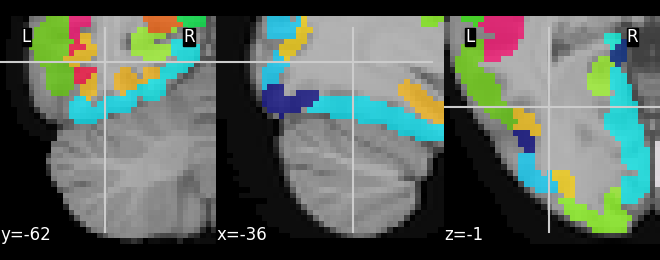
Regarding the resampling solution I tried to upload the image but the filetype is not supported, so I cannot share it. However I used the 700um probabilistic file from here OSF | Amygdala Atlas Files and adjusted it with a threshold of 0.7 and multiplied each dimension with 1,2,3… to distinguish the nuclei.
And the code to reproduce the error was kind of the same what you posted
img = image.resample_img(
atlas2, target_affine=atlas2.affine, target_shape=(40, 40, 40)
)
bg = image.load_img(datasets.load_mni152_template(2))
bg = image.resample_to_img(bg, img)
disp = plotting.plot_roi(img, bg_img=bg)
plotting.show()
Unfortunately now if have another problem with my plots as they do not save correctly (actually being plain white). Can I just ask you again if that is related to the issue of showing the image before saving as described here: Saving plots - Problem Solving with Python?
I tried it with %matplotlib auto to disable the inline plotting but same problem.
This is my code:
y_slices = [-15,-14,-13,-12,-11,-10,-9,-8,-7,-6,-5,-4,-3,-2,-1,0,1]
# set colors of the different nuclei
la = ListedColormap([9/255, 215/255, 200/255])
bldi = ListedColormap([9/255, 197/255, 147/255])
bm = ListedColormap([63/255, 195/255, 58/255])
ce = ListedColormap([245/255, 0/255, 0/255])
cmn = ListedColormap([130/255, 5/255, 90/255])
blpl = ListedColormap([113/255, 169/255, 140/255])
ata = ListedColormap([240/255, 80/255, 190/255])
asta = ListedColormap([250/255, 150/255, 150/255])
aaa = ListedColormap([255/255, 225/255, 0/255])
for y_slice in y_slices:
display = plotting.plot_roi(image.index_img(TP_img, 0), threshold=0.7, cmap=la, display_mode='y', cut_coords = [y_slice],
colorbar=False, axes=(100,100,10,10))
display.add_overlay(image.index_img(TP_img, 0),threshold=0.7,
cmap=la) # LA
display.add_overlay(image.index_img(TP_img, 1),threshold=0.7,
cmap=bldi) # BLDI
display.add_overlay(image.index_img(TP_img, 2),threshold=0.7,
cmap=bm) # BM
display.add_overlay(image.index_img(TP_img, 3),threshold=0.7,
cmap=ce) # CE
display.add_overlay(image.index_img(TP_img, 4),threshold=0.7,
cmap=cmn) # CMN
display.add_overlay(image.index_img(TP_img, 5),threshold=0.7,
cmap=blpl)
display.add_overlay(image.index_img(TP_img, 6),threshold=0.7,
cmap=ata) # ATA
display.add_overlay(image.index_img(TP_img, 7),threshold=0.7,
cmap=asta) # ASTA
display.add_overlay(image.index_img(TP_img, 8),threshold=0.7,
cmap=aaa) # AAA
for cut_ax in display.axes.values():
cut_ax.ax.set_xlim(-40, 40)
cut_ax.ax.set_ylim(-40, 40)
display.savefig(f'{plot_dir}Masks/Atlas/th70_{y_slice}.svg')
display.close()