Hi Everyone , I am working with .nii fMRI files . Is there any way to find out which particular frame has some information and which frames have no information. This is required to avoid sending empty frames to training network.
Hi @Deepika_Garg, and welcome to neurostars!
May you expand on what you mean by “has some information” please?
Best,
Steven
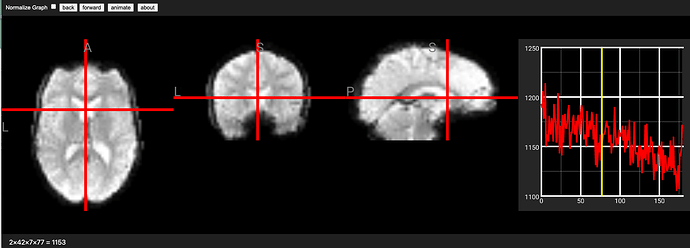
You can simply drag and drop your NIfTI image onto this NiiVue live demo. You can click on the 2D slices to select a voxel and click on the timeline to select a volume (you can also use the mouse scrollwheel while hovering over the timeline to select voxels). The volume number is shown as the 4th dimension in the status bar at the bottom right corner (e.g. the 77th volume). Be aware that most neuroimaging tools (nilearn, FSL, NiiVue) are indexed from 0, e.g. the first volume is volume zero, but Matlab-based tools like SPM are indexed from 1 (the first volume is volume one).
Hi @Steven : i am trying to use a neural network for classifying fMRI samples as positive or negative regarding some particular disorder . I can’t give the full fMRI as input as it requires too much hardware configurations and system time as well. I have augmented the fMRI images into smaller .nii files using slice function of nilearn. When i checked these sliced files using BrainViewer, there are many empty .nii files as there may be no activity during that time frame. It is causing the network training to go haywire.
I tried comparing 1 active and 1 empty .nii files as well using get_fdata() . But both images are showing similar matrices. Also It is not possible to check each and every file manually . Is there any way to check whether input .nii file is having some activations or not.
Also is there any good tool to visualize 4D fMRI data in windows .
Thanks.
Hi @neurolabusc : Each of my file is having >200 timestamps and more than 1000 such files.So it is not possible to check each timestamp individually .
Is there any function or matrix to know the same .
Thanks.