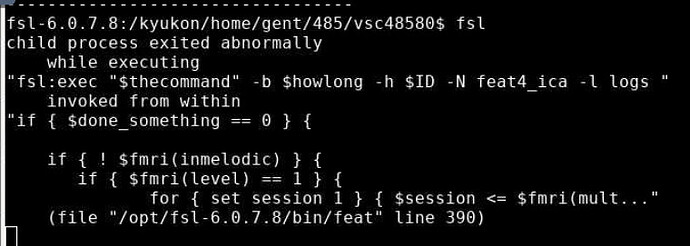
Hello! I am relatively new to the fMRI world and I am encountering some issues while computing some analyses on fsl. I have some .nifti resting state fMRI volumes, already pre-processed, that I want to use to retrieve intrinsic connectivity networks with fsl MELODIC. However, I get this error I attach in the image:
Hi @Ibai_Azpeitia_Loiti, the error messages emitted to the terminal by Feat and Melodic are typically not informative - can you look in the files contained within the <analysis>.feat/logs/ directory to see if there are any more useful error messages?
Hi @Ibai_Azpeitia_Loiti, that error suggests that there is something wrong with the input data to melodic - perhaps one of the preprocessing steps failed. Can you create a .zip file of your .feat directory and share it with me (using e.g. dropbox, google drive, etc)?
Hi @Ibai_Azpeitia_Loiti, I was hoping to look at the entire .feat directory, not just the .feat/logs directory.
Hi @paulmccarthy, you mean the whole .ica foler right?
https://www.dropbox.com/scl/fi/bg3rulrzcn01ialcr6v8l/Desktop.ica.zip?rlkey=gzn28tmo51kyp96wievhxz49a&st=ds6jc399&dl=0
Hi @Ibai_Azpeitia_Loiti, I don’t see any image files in your upload - I think that melodic is failing because of something in the input data, so I was hoping to look at the data to try and figure out why.
Hi @paulmccarthy this is one of the fMRI sessions I have been using:
https://www.dropbox.com/scl/fi/z6glb3ali13062n1nyp96/sub-cast1_ses-01_task-rest_bold_talairach.nii.gz?rlkey=d5380k6yvl2jip4qr4fiq84a3&st=qawx2rf1&dl=0
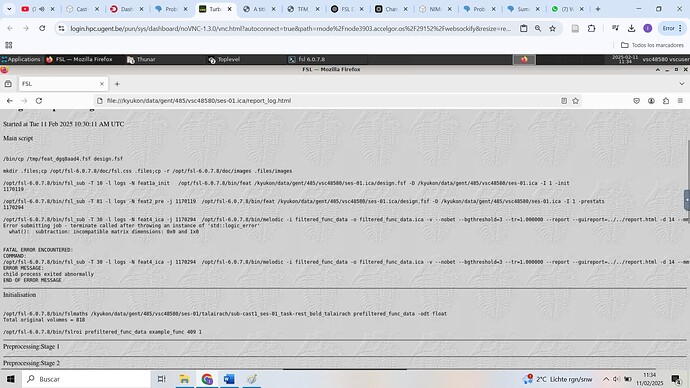
The file that you uploaded does not look like typical fMRI data - it has been heavily pre-processed and, more importantly, appears to have undergone some kind of intensity normalisation.
The standard MELODIC pipeline expects normal unprocessed or minimally preprocessed fMRI data, so you won’t be able to apply it to this kind of data - you will need to use the melodic command-line tool - I was able to get some results using this command:
melodic \
-i sub-cast1_ses-01_task-rest_bold_talairach.nii.gz \
-o output.ica \
--varnorm --nomask --keep_meanvol
For this data, the --keep_meanvol flag is necessary, as the input data appears to have already been de-meaned.
I would recommend that you take some more time to go through some educational material, and build a clearer picture of what you are trying to accomplish. There is plenty of material online for learning about neuroimaging research - for example, you could start by going through the FSL course (all of the lectures, data, and practicals are freely available): FSL Course