Hi all,
I’m looking for some advice regarding processing single-subject, non-whole brain functional images.
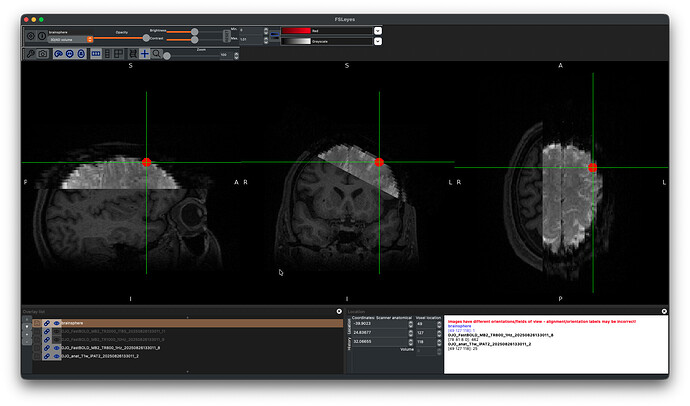
I ran an experiment to attempt to measure potential BOLD response to TMS at the site of stimulation. To optimize spatial resolution, we used an acquisition FOV limited to the region of interest, obliqued to cover just the scalp and a few inches into the brain underneath the TMS coil.
Because of this limited FOV, I wasn’t sure which standard processing steps would still apply or provide reasonably accurate information.
Here is a example of what I’m working with.
Ideally, we would like to perform a “task” analysis on the images to identify BOLD response within a sphere around the stim site (5mm red sphere).
We did one run of each of three stimulation paradigms, all with variable numbers of volumes and acquisition parameters.
In addition to the task images, we also acquired a reverse PE image to perform field map correction, as well as a long-TR whole brain image with the same rotational FOV for assistance in registration.
What is the best way to go about getting the cleanest signal before performing this analysis?
I have used fmriprep in the past, but after an initial pass on this data it seemed to struggle with aligning the slab images accurately to the T1 so I’m concerned about using any of the confound files to denoise the signal. I have also tried using FSL’s tools to manually realign/deoblique and fieldmap-correct the images before pushing them through the GLM, but again, I’m not super confident in the outputs based on the application of techniques and lack of denoising applied.
Any suggestions are greatly appreciated!
Thanks,
Jess