How to process nifti files in machine learning. when converting to other image formats resulting 120 slices.
What other image formats did you have in mind? There are many neuroimaging software packages that use machine learning classifiers and are compatible with nifti images.
When I converted nifti files to other image formats like jpg, png , I got 120 slices. Can you please tell me some neuroimaging software packages.
Thankyou for your response.
No need to convert outside of Nifti. Nilearn (https://nilearn.github.io/) is a popular python library for neuroscience processing.
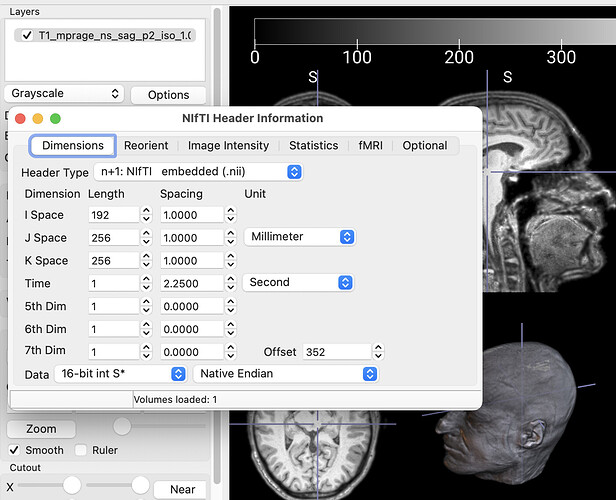
I agree that using a tool like Nilearn that is designed for NIfTI images is a wise approach. If you want more help, you may want to provide more details about your images. For example, the command line tool fslhd can report details, as can the graphical MRIcroGL program. The image below shows the result of choosing the Show Header menu item from the Options button. In this example, note that like most medical images, this image is saved with 16-bit signed precision: voxels can have 65535 values of gray. In contrast, typical 24-bit RGB or 32-bit RGBA JPEG and PNG images are restricted to just 256 levels of gray. Therefore, if you wish to export the data, you must set the window center and width (brightness and contrast) to restrict the dynamic range of your image. This will likely impact the accuracy of your subsequent analyses. The goal is to have solutions that are not merely better than chance, but accurate enough to change the standard of care. Preserving the precision of your data helps in this aim.
thank you so much for your information.
Thank you for your information.
It’s very useful. I am using the dataset ( Anatomical Tracings of Lesions After Stroke (ATLAS ) R1.2. Do you have any idea about this.
What kind of information are you looking for?
Need to process ischemic stroke brain mri images in machine learning.
To classify normal and abnormal. I’ve just started with nilearn. Thank you for your valuable information.