Hi again Julia,
The streamlines generated by tractography algorithms don’t usually inherently have a particular direction that they are organized in. So, for example, two neighboring streamlines in a tractography may be going in opposite directions, with one that has, for example, coordinates that are more anterior followed by more posterior coordinates and the other could have coordinates that are more posterior followed by more anterior coordinates. In pyAFQ, we make sure that all the streamlines within one bundle are oriented in the same way by checking whether each streamline goes first through the first waypoint ROI in the bundle definition and flipping its coordinates if it doesn’t. So, when you observe that some bundles in pyAFQ are organized from posterior to anterior that’s because in the bundle definitions, we usually have the posterior ROI first (but you are correct that’s not always the case!). We do intend to provide a more complete description of each bundle’s orientation in the documentation, but have not done so yet.
In the DIPY example, a similar goal is achieved through this code:
qb = QuickBundles(np.inf, metric=metric)
cluster_cst_l = qb.cluster(model_cst_l)
standard_cst_l = cluster_cst_l.centroids[0]
oriented_cst_l = dts.orient_by_streamline(cst_l, standard_cst_l)
Which makes sure that all the streamlines are oriented in the same way as the centroid of the clustering procedure. However, this clustering procedure can generate a centroid that is oriented in either direction and different orientations on different runs. So, for your bundles, you’d want to figure out a way to orient your streamlines consistently across runs and across individuals. Because this depends a bit on the overall orientation of the bundle, there isn’t a catch-all solution for this, though (except to use pyAFQ whenever possible  ).
).
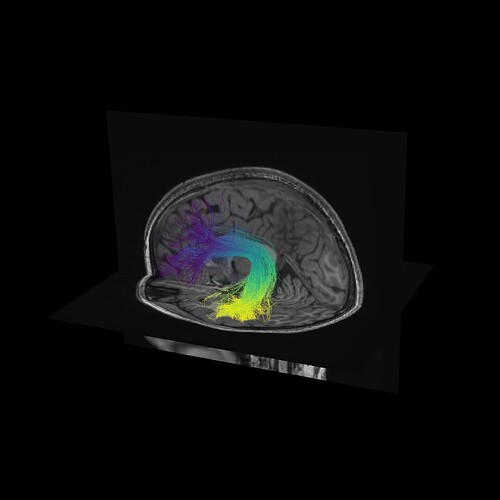
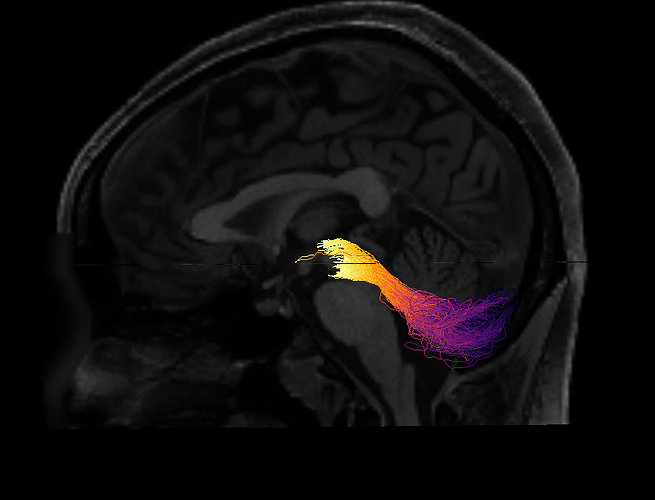
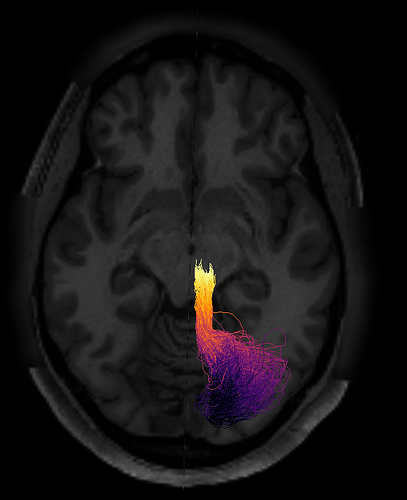
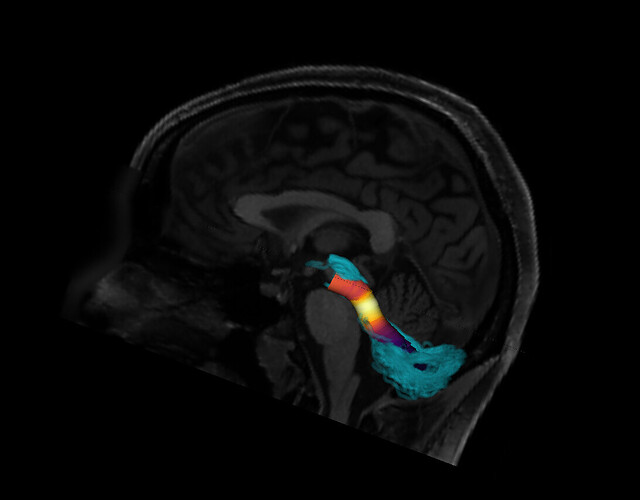
One way to verify the orientation of a particular tract profile is to visualize it. I would create a synthetic tract profile that has the numbers [0, 1, 2,...,99] in it and then plot it colorizing each of the streamlines in the bundle with these numbers. What you should see is something like this:
This was generated using the following code (which should be self-contained and downloads the data it needs):
import os.path as op
import nibabel as nib
import numpy as np
from dipy.io.streamline import load_trk
from dipy.tracking.streamline import transform_streamlines
from fury import actor, window
from fury.colormap import create_colormap
import AFQ.data.fetch as afd
afd.fetch_hbn_preproc(["NDARAA948VFH"])
study_path = afd.fetch_hbn_afq(["NDARAA948VFH"])[1]
deriv_path = op.join(
study_path, "derivatives")
afq_path = op.join(
deriv_path,
'afq',
'sub-NDARAA948VFH',
'ses-HBNsiteRU')
bundle_path = op.join(afq_path,
'bundles')
fa_img = nib.load(op.join(afq_path,
'sub-NDARAA948VFH_ses-HBNsiteRU_acq-64dir_space-T1w_desc-preproc_dwi_model-DKI_FA.nii.gz'))
fa = fa_img.get_fdata()
sft_arc = load_trk(op.join(bundle_path,
'sub-NDARAA948VFH_ses-HBNsiteRU_acq-64dir_space-T1w_desc-preproc_dwi_space-RASMM_model-CSD_desc-prob-afq-ARC_L_tractography.trk'), fa_img)
sft_cst = load_trk(op.join(bundle_path,
'sub-NDARAA948VFH_ses-HBNsiteRU_acq-64dir_space-T1w_desc-preproc_dwi_space-RASMM_model-CSD_desc-prob-afq-CST_L_tractography.trk'), fa_img)
t1w_img = nib.load(op.join(deriv_path,
'qsiprep/sub-NDARAA948VFH/anat/sub-NDARAA948VFH_desc-preproc_T1w.nii.gz'))
t1w = t1w_img.get_fdata()
sft_arc.to_rasmm()
sft_cst.to_rasmm()
arc_t1w = transform_streamlines(sft_arc.streamlines,
np.linalg.inv(t1w_img.affine))
cst_t1w = transform_streamlines(sft_cst.streamlines,
np.linalg.inv(t1w_img.affine))
def lines_as_tubes(sl, line_width, **kwargs):
line_actor = actor.line(sl, **kwargs)
line_actor.GetProperty().SetRenderLinesAsTubes(1)
line_actor.GetProperty().SetLineWidth(line_width)
return line_actor
def slice_volume(data, x=None, y=None, z=None):
slicer_actors = []
slicer_actor_z = actor.slicer(data)
if z is not None:
slicer_actor_z.display_extent(
0, data.shape[0] - 1,
0, data.shape[1] - 1,
z, z)
slicer_actors.append(slicer_actor_z)
if y is not None:
slicer_actor_y = slicer_actor_z.copy()
slicer_actor_y.display_extent(
0, data.shape[0] - 1,
y, y,
0, data.shape[2] - 1)
slicer_actors.append(slicer_actor_y)
if x is not None:
slicer_actor_x = slicer_actor_z.copy()
slicer_actor_x.display_extent(
x, x,
0, data.shape[1] - 1,
0, data.shape[2] - 1)
slicer_actors.append(slicer_actor_x)
return slicer_actors
slicers = slice_volume(t1w, x=t1w.shape[0] // 2, z=t1w.shape[-1] // 3)
scene = window.Scene()
scene.set_camera(position=(238.04, 174.48, 143.04),
focal_point=(96.32, 110.34, 84.48),
view_up=(-0.33, -0.12, 0.94))
sft_arc.to_vox()
scene.clear()
sl = list(arc_t1w)
for sl in arc_t1w:
arc_actor = actor.line(
[sl],
colors=create_colormap(np.linspace(0, 100, len(sl)), 'viridis'))
scene.add(arc_actor)
for slicer in slicers:
scene.add(slicer)
window.record(scene, out_path='colored_by_direction.png', size=(2400, 2400))