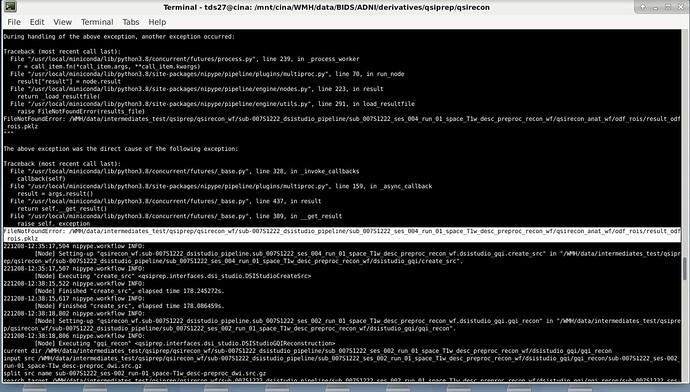
Yes, below is the error copied from the terminal.
[Node] Error on "qsirecon_wf.sub-007S1222_amico_noddi.sub_007S1222_ses_004_run_01_space_T1w_desc_preproc_recon_wf.qsirecon_anat_wf.resample_mask" (/WMH/data/intermediates/qsiprep/qsirecon_wf/sub-007S1222_amico_noddi/sub_007S1222_ses_004_run_01_space_T1w_desc_preproc_recon_wf/qsirecon_anat_wf/resample_mask)
exception calling callback for <Future at 0x7f759c9eeb20 state=finished raised FileNotFoundError>
concurrent.futures.process._RemoteTraceback:
"""
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 722, in _run_command
result = self._interface.run(cwd=outdir, ignore_exception=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 388, in run
self._check_mandatory_inputs()
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 275, in _check_mandatory_inputs
raise ValueError(msg)
ValueError: Resample requires a value for input 'in_file'. For a list of required inputs, see Resample.help()
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/concurrent/futures/process.py", line 239, in _process_worker
r = call_item.fn(*call_item.args, **call_item.kwargs)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 70, in run_node
result["result"] = node.result
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 223, in result
return _load_resultfile(
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/utils.py", line 291, in load_resultfile
raise FileNotFoundError(results_file)
FileNotFoundError: /WMH/data/intermediates/qsiprep/qsirecon_wf/sub-007S1222_amico_noddi/sub_007S1222_ses_004_run_01_space_T1w_desc_preproc_recon_wf/qsirecon_anat_wf/resample_mask/result_resample_mask.pklz
"""
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/concurrent/futures/_base.py", line 328, in _invoke_callbacks
callback(self)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 159, in _async_callback
result = args.result()
File "/usr/local/miniconda/lib/python3.8/concurrent/futures/_base.py", line 437, in result
return self.__get_result()
File "/usr/local/miniconda/lib/python3.8/concurrent/futures/_base.py", line 389, in __get_result
raise self._exception
FileNotFoundError: /WMH/data/intermediates/qsiprep/qsirecon_wf/sub-007S1222_amico_noddi/sub_007S1222_ses_004_run_01_space_T1w_desc_preproc_recon_wf/qsirecon_anat_wf/resample_mask/result_resample_mask.pklz
221209-13:10:54,578 nipype.workflow INFO:
[Node] Executing "resample_mask" <nipype.interfaces.afni.utils.Resample>
221209-13:10:54,737 nipype.workflow INFO:
[Node] Executing "odf_rois" <nipype.interfaces.ants.resampling.ApplyTransforms>
221209-13:10:54,738 nipype.workflow WARNING:
[Node] Error on "qsirecon_wf.sub-007S1222_amico_noddi.sub_007S1222_ses_004_run_01_space_T1w_desc_preproc_recon_wf.qsirecon_anat_wf.odf_rois" (/WMH/data/intermediates/qsiprep/qsirecon_wf/sub-007S1222_amico_noddi/sub_007S1222_ses_004_run_01_space_T1w_desc_preproc_recon_wf/qsirecon_anat_wf/odf_rois)
exception calling callback for <Future at 0x7f759c9a99a0 state=finished raised FileNotFoundError>
concurrent.futures.process._RemoteTraceback:
"""
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 722, in _run_command
result = self._interface.run(cwd=outdir, ignore_exception=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 388, in run
self._check_mandatory_inputs()
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 275, in _check_mandatory_inputs
raise ValueError(msg)
ValueError: ApplyTransforms requires a value for input 'transforms'. For a list of required inputs, see ApplyTransforms.help()