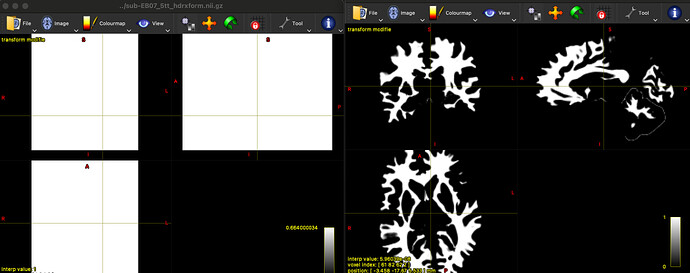
Summary of what happened:
Hi Qsirecon Exports,
I am using qsirecon-1.0.0rc1 to analyze preprocessed DTI data (from qsiprep-1.0.0rc1 ). The freesurfer data from fmriprep analysis. I have a problem, tckgen: Processing mask is empty; check input images / registration.
More importantly, I found this error only occurred in 2/30 subjects,and the other subjects can smoothly completed. So I am not sure what caused this question. And How should I solve this question.
Looking forward to your reply,
Sincerely
Command used (and if a helper script was used, a link to the helper script or the command generated):
apptainer run \
--containall \
--writable-tmpfs \
-B "${inputpath}":"${inputpath}" \
-B "${outputpath}":"${outputpath}" \
-B "${scratch_dir}":"${scratch_dir}" \
-B "${freesurfer_license_folder}":/opt/freesurfer/license.txt \
-B "${fsinput}":"${fsinput}" \
${path_to_singularity_image} \
"${inputpath}" \
"${outputpath}" \
participant \
--fs-license-file /opt/freesurfer/license.txt \
-w "${scratch_dir}" \
--fs-subjects-dir $fsinput \
--recon-spec mrtrix_singleshell_ss3t_ACT-hsvs \
--atlases 4S156Parcels 4S256Parcels Brainnetome246Ext Gordon333Ext AAL116 \
--output-resolution 1.2 \
-v -v \
--stop-on-first-crash \
--participant-label $subjID
Version:
1.0.0rc1
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 66, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 525, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 643, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 769, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node tractography.
Cmdline:
tckgen -act /globalscratch/ucl/irsp/yufenghe/work-qsirecon/sub-EB07/qsirecon_1_0_wf/sub-EB07_mrtrix_singleshell_ss3_hsvst/recon_anatomical_wf_0/apply_header_to_5tt/sub-EB07_5tt_hdrxform.nii.gz -algorithm iFOD2 -backtrack -crop_at_gmwmi -maxlength 250.000000 -minlength 30.000000 -samples 4 -nthreads 8 -output_seeds out_seeds.nii.gz -power 0.330000 -quiet -seed_dynamic /globalscratch/ucl/irsp/yufenghe/work-qsirecon/sub-EB07/qsirecon_1_0_wf/sub-EB07_mrtrix_singleshell_ss3_hsvst/sub_EB07_ses_01_space_ACPC_desc_preproc_recon_wf/ss3t_csd/intensity_norm/sub-EB07_ses-01_space-ACPC_desc-preproc_dwi_wm_mtnorm.mif -select 10000000 /globalscratch/ucl/irsp/yufenghe/work-qsirecon/sub-EB07/qsirecon_1_0_wf/sub-EB07_mrtrix_singleshell_ss3_hsvst/sub_EB07_ses_01_space_ACPC_desc_preproc_recon_wf/ss3t_csd/intensity_norm/sub-EB07_ses-01_space-ACPC_desc-preproc_dwi_wm_mtnorm.mif tracked.tck
Stdout:
Stderr:
tckgen: e[01;31m[ERROR] Processing mask is empty; check input images / registratione[0m
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 457, in aggregate_outputs
setattr(outputs, key, val)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 325, in validate
value = super().validate(objekt, name, value, return_pathlike=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: The 'out_file' trait of a TractographyOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/globalscratch/ucl/irsp/yufenghe/work-qsirecon/sub-EB07/qsirecon_1_0_wf/sub-EB07_mrtrix_singleshell_ss3_hsvst/sub_EB07_ses_01_space_ACPC_desc_preproc_recon_wf/track_ifod2/tractography/tracked.tck' <class 'str'> was specified.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 404, in run
outputs = self.aggregate_outputs(runtime)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 464, in aggregate_outputs
raise FileNotFoundError(msg)
FileNotFoundError: No such file or directory '/globalscratch/ucl/irsp/yufenghe/work-qsirecon/sub-EB07/qsirecon_1_0_wf/sub-EB07_mrtrix_singleshell_ss3_hsvst/sub_EB07_ses_01_space_ACPC_desc_preproc_recon_wf/track_ifod2/tractography/tracked.tck' for output 'out_file' of a TckGen interface