screen docker run -ti \
-v /Users/neurouser1/Documents/ENRICH/qsiprepout_controls/qsiprep/:/data \
-v /Users/neurouser1/Documents/ENRICH/qsiprepmrtrix_controls/:/out \
-v /Users/neurouser1/Documents/fs_files/:/license \
-v /Users/neurouser1/Documents/ENRICH/scratch/:/scratch \
-v /Users/neurouser1/Documents/ENRICH/xcpd_controls/fmriprep_out/sourcedata/freesurfer/:/freesurferdir \
pennlinc/qsirecon:0.23.2 \
/data /out participant \
--participant_label sub-ENRH04 \
--output-resolution 1.3 \
--n-cpus 15 \
--omp-nthreads 8 \
--mem-mb 150000 \
--fs-license-file /license/license.txt \
--recon-spec mrtrix_multishell_msmt_ACT-hsvs \
--freesurfer-input /freesurferdir/ \
-w /scratch/ \
--resource-monitor
pennlinc/qsirecon:0.23.2
Docker container
Yes
250627-21:38:48,989 nipype.workflow INFO:
[Node] Setting-up "qsirecon_0_23_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.estimate_fod" in "/scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/estimate_fod".
250627-21:38:49,20 nipype.workflow INFO:
[Node] Executing "estimate_fod" <qsirecon.interfaces.mrtrix.EstimateFOD>
250627-22:08:18,541 nipype.workflow INFO:
[Node] Finished "estimate_fod", elapsed time 1769.520133s.
250627-22:08:18,939 nipype.workflow INFO:
[Node] Setting-up "qsirecon_0_23_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.intensity_norm" in "/scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/intensity_norm".
250627-22:08:18,969 nipype.workflow INFO:
[Node] Executing "intensity_norm" <qsirecon.interfaces.mrtrix.MTNormalize>
250627-22:08:51,389 nipype.workflow INFO:
[Node] Finished "intensity_norm", elapsed time 32.419709s.
250627-22:08:53,797 nipype.workflow INFO:
[Node] Setting-up "qsirecon_0_23_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.plot_peaks" in "/scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/plot_peaks".
250627-22:08:54,219 nipype.workflow INFO:
[Node] Executing "plot_peaks" <qsirecon.interfaces.reports.CLIReconPeaksReport>
250627-22:10:35,974 nipype.workflow INFO:
[Node] Finished "plot_peaks", elapsed time 101.650972s.
250627-22:10:36,734 nipype.workflow INFO:
[Node] Setting-up "qsirecon_0_23_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.track_ifod2.tractography" in "/scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/track_ifod2/tractography".
250627-22:10:36,764 nipype.workflow INFO:
[Node] Executing "tractography" <qsirecon.interfaces.mrtrix.TckGen>
I have already used this qsirecon version on 3 other subjects and they took around 5 hours to run and completed successfully.
Any insight is appreciated.
Thank you
Here are the crash logs in case they are helpful.
Crash log 1
Node: qsirecon_0_23_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.recon_anatomical_wf.ds_fs_5tt_hsvs
Working directory: /scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/recon_anatomical_wf/ds_fs_5tt_hsvs
Node inputs:
acquisition = <undefined>
atlas = <undefined>
base_directory = /out
bundle = <undefined>
bundles = <undefined>
ceagent = <undefined>
check_hdr = True
chunk = <undefined>
cohort = <undefined>
compress = [True]
data_dtype = <undefined>
datatype = <undefined>
den = <undefined>
desc = hsvs
direction = <undefined>
dismiss_entities = ['desc']
echo = <undefined>
extension = <undefined>
flip = <undefined>
fmap = <undefined>
from = <undefined>
hemi = <undefined>
in_file = ['/scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/recon_anatomical_wf/create_5tt_hsvs/sub-ENRH04_5tt.mif']
inv = <undefined>
label = <undefined>
meta_dict = <undefined>
modality = <undefined>
mode = <undefined>
model = <undefined>
mt = <undefined>
nucleus = <undefined>
param = <undefined>
part = <undefined>
proc = <undefined>
reconstruction = <undefined>
recording = <undefined>
res = <undefined>
roi = <undefined>
run = <undefined>
sample = <undefined>
scans = <undefined>
session = <undefined>
source_file = ['/data/sub-ENRH04/anat/sub-ENRH04_desc-preproc_T1w.nii.gz']
space = fsnative
staining = <undefined>
subject = <undefined>
subset = <undefined>
suffix = dseg
task = <undefined>
to = <undefined>
tracer = <undefined>
tracksys = <undefined>
volume = <undefined>
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 344, in _send_procs_to_workers
self.procs[jobid].run(updatehash=updatehash)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node ds_fs_5tt_hsvs.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/niworkflows/interfaces/bids.py", line 632, in _run_interface
raise ValueError(f"Could not build path with entities {out_entities}.")
ValueError: Could not build path with entities {'subject': 'ENRH04', 'suffix': 'dseg', 'datatype': 'anat', 'extension': 'mif.gz', 'desc': 'hsvs', 'space': 'fsnative'}.
Crash log 2:
Node: qsirecon_0_23_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.recon_anatomical_wf.ds_qsiprep_5tt_hsvs
Working directory: /scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/recon_anatomical_wf/ds_qsiprep_5tt_hsvs
Node inputs:
acquisition = <undefined>
atlas = hsvs
base_directory = /out
bundle = <undefined>
bundles = <undefined>
ceagent = <undefined>
check_hdr = True
chunk = <undefined>
cohort = <undefined>
compress = <undefined>
data_dtype = <undefined>
datatype = <undefined>
den = <undefined>
desc = <undefined>
direction = <undefined>
dismiss_entities = ['desc']
echo = <undefined>
extension = <undefined>
flip = <undefined>
fmap = <undefined>
from = <undefined>
hemi = <undefined>
in_file = ['/scratch/qsirecon_0_23_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/recon_anatomical_wf/apply_header_to_5tt/sub-ENRH04_5tt_hdrxform.nii.gz']
inv = <undefined>
label = <undefined>
meta_dict = <undefined>
modality = <undefined>
mode = <undefined>
model = <undefined>
mt = <undefined>
nucleus = <undefined>
param = <undefined>
part = <undefined>
proc = <undefined>
reconstruction = <undefined>
recording = <undefined>
res = <undefined>
roi = <undefined>
run = <undefined>
sample = <undefined>
scans = <undefined>
session = <undefined>
source_file = ['/data/sub-ENRH04/anat/sub-ENRH04_desc-preproc_T1w.nii.gz']
space = T1w
staining = <undefined>
subject = <undefined>
subset = <undefined>
suffix = dseg
task = <undefined>
to = <undefined>
tracer = <undefined>
tracksys = <undefined>
volume = <undefined>
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 344, in _send_procs_to_workers
self.procs[jobid].run(updatehash=updatehash)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node ds_qsiprep_5tt_hsvs.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/niworkflows/interfaces/bids.py", line 632, in _run_interface
raise ValueError(f"Could not build path with entities {out_entities}.")
ValueError: Could not build path with entities {'subject': 'ENRH04', 'suffix': 'dseg', 'datatype': 'anat', 'extension': 'nii', 'atlas': 'hsvs', 'space': 'T1w'}.
Thank you
tsalo
June 30, 2025, 6:56pm
3
Can you updating to a newer version (e.g., 1.1.0)? I think that particular bug should be fixed in the newest release.
Also, I’d recommend including --stop-on-first-crash. That should hopefully stop the workflow from stalling.
Hi @tsalo thank you so much for the recommendation.
I tried running qsirecon same command as above with verson 1.1.0. (and the --stop-on-first-crash flag). The 2 errors are gone and no crash reports are generated but the workflow has still stalled on tckgen. The docker container is still running for 22 hours. Any other suggestions on how to resolve this?
Thank you
[Node] Executing "apply_header_to_5tt" <qsirecon.interfaces.mrtrix.TransformHeader>
250630-21:15:37,765 nipype.workflow INFO:
[Node] Finished "apply_header_to_5tt", elapsed time 0.067126s.
250630-21:15:39,705 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_inputs" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_inputs".
250630-21:15:40,396 nipype.workflow INFO:
[Node] Executing "sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_inputs" <qsirecon.interfaces.interchange.ReconWorkflowInputs>
250630-21:15:40,397 nipype.workflow INFO:
[Node] Finished "sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_inputs", elapsed time 0.000539s.
250630-21:15:41,306 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.create_mif" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/create_mif".
250630-21:15:41,352 nipype.workflow INFO:
[Node] Executing "create_mif" <qsirecon.interfaces.mrtrix.MRTrixIngress>
250630-21:15:48,309 nipype.interface INFO:
mrconvert: [100%] uncompressing image "/data/sub-ENRH04/ses-Baseline/dwi/sub-ENRH04_ses-Baseline_space-T1w_desc-preproc_dwi.nii.gz"
250630-21:15:54,587 nipype.interface INFO:
mrconvert: [100%] copying from "/data/sub-...T1w_desc-preproc_dwi.nii.gz" to "/scratch/q...ce-T1w_desc-preproc_dwi.mif"
250630-21:16:01,244 nipype.workflow INFO:
[Node] Finished "create_mif", elapsed time 19.891747s.
250630-21:16:03,327 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.estimate_response" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/estimate_response".
250630-21:16:03,368 nipype.workflow INFO:
[Node] Executing "estimate_response" <qsirecon.interfaces.mrtrix.SS3TDwi2Response>
250630-21:17:24,193 nipype.workflow INFO:
[Node] Finished "estimate_response", elapsed time 80.824521s.
250630-21:17:25,351 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.estimate_fod" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/estimate_fod".
250630-21:17:25,395 nipype.workflow INFO:
[Node] Executing "estimate_fod" <qsirecon.interfaces.mrtrix.EstimateFOD>
250630-21:46:24,921 nipype.workflow INFO:
[Node] Finished "estimate_fod", elapsed time 1739.458896s.
250630-21:46:26,531 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.intensity_norm" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/intensity_norm".
250630-21:46:26,574 nipype.workflow INFO:
[Node] Executing "intensity_norm" <qsirecon.interfaces.mrtrix.MTNormalize>
250630-21:46:57,782 nipype.workflow INFO:
[Node] Finished "intensity_norm", elapsed time 31.138846s.
250630-21:46:59,123 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.msmt_csd.plot_peaks" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/msmt_csd/plot_peaks".
250630-21:46:59,164 nipype.workflow INFO:
[Node] Executing "plot_peaks" <qsirecon.interfaces.reports.CLIReconPeaksReport>
250630-21:48:39,218 nipype.workflow INFO:
[Node] Finished "plot_peaks", elapsed time 100.05341s.
250630-21:48:40,896 nipype.workflow INFO:
[Node] Setting-up "qsirecon_1_1_wf.sub-ENRH04_mrtrix_multishell_msmt_hsvs.sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf.track_ifod2.tractography" in "/scratch/qsirecon_1_1_wf/sub-ENRH04_mrtrix_multishell_msmt_hsvs/sub_ENRH04_ses_Baseline_space_T1w_desc_preproc_recon_wf/track_ifod2/tractography".
250630-21:48:40,939 nipype.workflow INFO:
[Node] Executing "tractography" <qsirecon.interfaces.mrtrix.TckGen>
Hi @apoorvakelkar ,
Whole brain tractography takes a while, what’s the longest you’ve let it run?
Best,
Hi @Steven its been running for 43 hours now and still no output. Typically, it’s taken around 12-16 hours per subject on our machine.
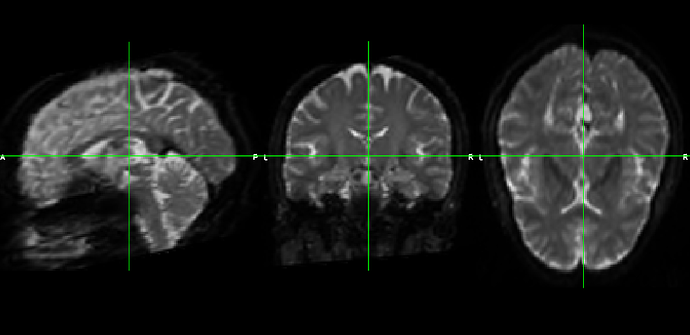
We did find the qsiprep preprocessed output dwi scan still has noise in it and looks grainy. Do you think that is why it’s taking a long time to compute the tractography? Your insight is appreciated.
Preprocessed output:
Thank you