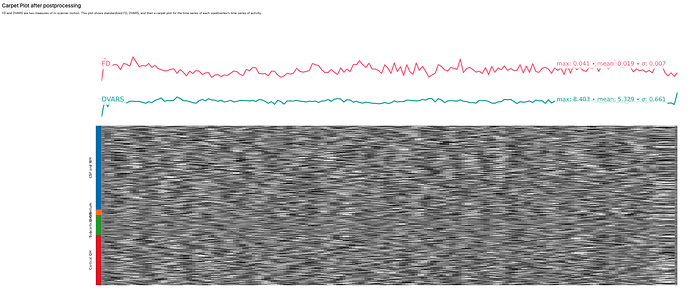
I spoke too soon.  xcp_d worked very well for a single task, but when I ran my first subject, I had several crash reports. They were all essentially the same, and only related to the node xcpd_wf.single_subject_400_wf.nifti_postprocess_2_wf.execsummary_wf.plot_svgx_wf. It seems that everything else ran just fine - there is just no Executive Summary .html. There is not really a pattern to which tasks led to the error - they were pretty evenly distributed across sessions, and different tasks for each session (e.g. ses-1: movie, rest-run2; ses-2: inscapes_run2, rest_run2, reward).
xcp_d worked very well for a single task, but when I ran my first subject, I had several crash reports. They were all essentially the same, and only related to the node xcpd_wf.single_subject_400_wf.nifti_postprocess_2_wf.execsummary_wf.plot_svgx_wf. It seems that everything else ran just fine - there is just no Executive Summary .html. There is not really a pattern to which tasks led to the error - they were pretty evenly distributed across sessions, and different tasks for each session (e.g. ses-1: movie, rest-run2; ses-2: inscapes_run2, rest_run2, reward).
EDITED TO ADD: I figured out what is going on. The exec summary node runs fine when there is no censoring. However, when there is censoring, the “regressed_data” input does not match the length of “raw_data” or “filtered_motion”, and thus gives the error. Please let me know if I should post on github! Thank you!
Additional info below…
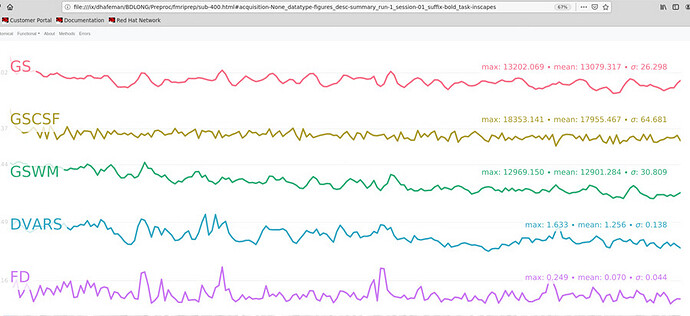
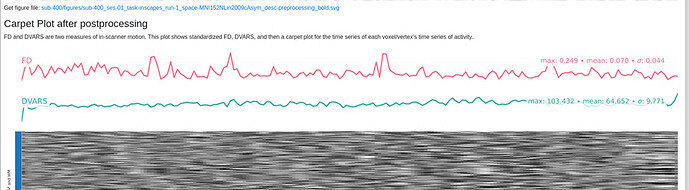
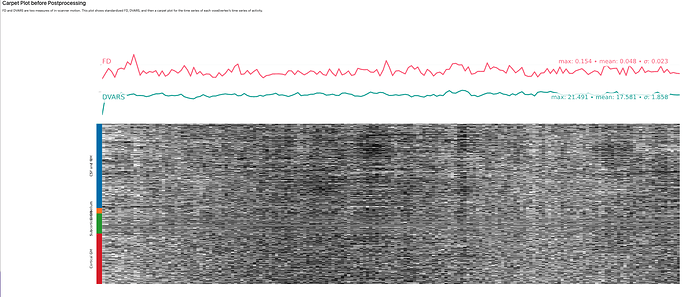
Here is the crash message. I would like to sort this out before running all of my subjects, because I do find the Executive Summary quite helpful.
Node: xcpd_wf.single_subject_400_wf.nifti_postprocess_2_wf.execsummary_wf.plot_svgx_wf
Working directory: /home/xcp_d/BDLONG/Work/xcpd_wf/single_subject_400_wf/nifti_postprocess_2_wf/execsummary_wf/plot_svgx_wf
Node inputs:
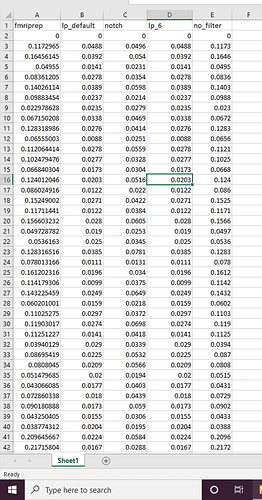
TR = 1.5
filtered_motion =
mask = /home/xcp_d/BDLONG/Preproc/fmriprep/sub-400/ses-01/func/sub-400_ses-01_task-movie_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
rawdata = /home/xcp_d/BDLONG/Preproc/fmriprep/sub-400/ses-01/func/sub-400_ses-01_task-movie_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz
regressed_data =
residual_data =
seg_data =
Traceback (most recent call last):
File “/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py”, line 67, in run_node
result[“result”] = node.run(updatehash=updatehash)
File “/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py”, line 527, in run
result = self._run_interface(execute=True)
File “/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py”, line 645, in _run_interface
return self._run_command(execute)
File “/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py”, line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node plot_svgx_wf.
Traceback:
Traceback (most recent call last):
File “/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py”, line 398, in run
runtime = self._run_interface(runtime)
File “/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/interfaces/surfplotting.py”, line 206, in _run_interface
‘after_process’] = plot_svgx(
File “/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/utils/plot.py”, line 523, in plot_svgx
DVARS_timeseries = pd.DataFrame({
File “/usr/local/miniconda/lib/python3.8/site-packages/pandas/core/frame.py”, line 529, in init
mgr = init_dict(data, index, columns, dtype=dtype)
File “/usr/local/miniconda/lib/python3.8/site-packages/pandas/core/internals/construction.py”, line 287, in init_dict
return arrays_to_mgr(arrays, data_names, index, columns, dtype=dtype)
File “/usr/local/miniconda/lib/python3.8/site-packages/pandas/core/internals/construction.py”, line 80, in arrays_to_mgr
index = extract_index(arrays)
File “/usr/local/miniconda/lib/python3.8/site-packages/pandas/core/internals/construction.py”, line 401, in extract_index
raise ValueError(“arrays must all be same length”)
ValueError: arrays must all be same length
Also, in case it is helpful, here was the code that I ran on our computing cluster:
singularity run --cleanenv -B /ix/dhafeman:/home/xcp_d -B /ix/dhafeman/license.txt:/opt/freesurfer/license.txt /ix/dhafeman/singularity/xcp_d-0.2.0.simg /home/xcp_d/BDLONG/Preproc/fmriprep /home/xcp_d/BDLONG/ participant -f 0.1 -w /home/xcp_d/BDLONG/Work --smoothing 8 --despike -p 36P --lower-bpf .009 --upper-bpf .08 --motion-filter-type lp --band-stop-min 6 --participant_label 400
Please also let me know if you would like me to post this as a new issue on neurostars or github. Thank you in advance for your help!