Hi everyone,
I am new to nilearn (and to python in general). I have been trying out different Nilearn functions to de-noise 7T resting state data, that was preprocessed using fmriprep. I tried nilearn.image.clean_img, nilearn.signal.clean and nilearn.maskers.NiftiMasker but each time the resultant de-noised image looks weird.
I copied and pasted a brief example of the code I used for each function below, along with an example output image. I would really appreciate it if someone can tell me what I am doing wrong / what might be going wrong and how I can fix it.
Thank you in advance.
–
from nilearn import image as nimg
from nilearn.maskers import NiftiMasker
import numpy as np
import pandas as pd
mask_file='/path/to/the/mni_normalized/mask/subj_mask_normalized.nii.gz'
mask_img = nimg.load_img(mask_file)
func_file='/path/to/the/mni_normalized/func/subj_normalized_bold.nii.gz'
func_img = nimg.load_img(func_file)
confound_file='/path/to/the/confound/file/confounds.tsv'
confounds= pd.read_csv("confounds.tsv", delimiter ='\t')
confounds=confounds.values
#(confounds include motion, WM, CSF, and derivatives for all from fmriprep, RETROICOR regressors derived using a toolbox)
image.clean_img
clean_img= nimg.clean_img(func_img, confounds=confounds, detrend=True, standardize=False, low_pass=0.2, high_pass=0.01, t_r=0.8, mask_img=mask_img)
clean_img.to_filename('clean_img_denoised_bold.nii.gz')
Output:
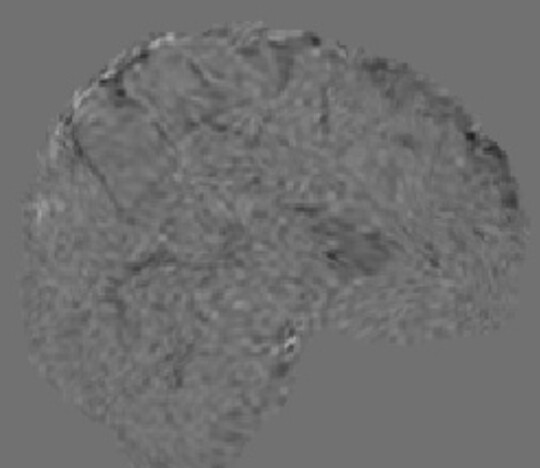
signal.clean
func_data=func_img.get_fdata()
func_data = np.reshape(func_data, (Number_of_Trs, -1)) #matching signal.shape[0] and confound.shape[0]
signal_cleaned=nilearn.signal.clean(func_data, runs=None, detrend=True, standardize=False, sample_mask=None, confounds=confounds, standardize_confounds=False, filter='butterworth', low_pass=0.2, high_pass=0.01, t_r=0.8, ensure_finite=False)
clean_data = np.reshape(signal_cleaned, func_img.shape)
clean_img= nb.Nifti1Image(clean_data, func_img.get_affine(),
func_img.get_header())
clean_img.to_filename('signal_clean_denoised_bold.nii.gz')
The output from this one looks more like a normal functional image, but the image has stripes on the it and it seems have a background added to it now (the input image here was skull-stripped with no background).
Niftimasker
#based on the whole brain time series extraction given in the example https://nilearn.github.io/stable/auto_examples/03_connectivity/plot_seed_to_voxel_correlation.html#sphx-glr-auto-examples-03-connectivity-plot-seed-to-voxel-correlation-py
brain_masker=NiftiMasker(mask_img=mask_img, runs=None, smoothing_fwhm=3.2, standardize=False, standardize_confounds=False, detrend=False, high_variance_confounds=False, low_pass=0.2, high_pass=0.01, t_r=0.8, memory_level=1, memory='nilearn cache', verbose=0, reports=True)
brain_time_series=brain_masker.fit_transform(func_img, confounds)
inversed=brain_masker.inverse_transform(brain_time_series)
inversed.to_filename('masker_denoised_bold.nii.gz')
Output image here looks a bit more like above, but better (still nothing like a functional image).
Regarding NiftiMasker
#When I initiated the fit_transform, I got this warning;
*Generation of a mask has been requested (y != None) while a mask has been provided at masker creation. Given mask will be used. warnings.warn('[%s.fit] Generation of a mask has been'*
#And this one below after it ran
*local/lib/python3.9/site-packages/nilearn/maskers/nifti_masker.py:570: UserWarning: Persisting input arguments took 31.77s to run.* If this happens often in your code, it can cause performance problems (results will be correct in all cases).
*The reason for this is probably some large input arguments for a wrapped function (e.g. large strings). THIS IS A JOBLIB ISSUE. If you can, kindly provide the joblib's team with an example so that they can fix the problem. data = self._cache(*