First of all, thanks to everyone involved in developing and maintaining tedana, it’s seems like a great community-driven project.
I’m running into some issues with tedana v0.0.9 (released on February 5th 2021) though and would therefore like to run tedana in debug mode. However, in contrast to earlier versions, the packaging of this release somehow has made it more difficult for me to figure out how to do this.
Could someone provide a few pointers on how to run tedana in debug mode (preferably using VS code)?
Thanks in advance!
Additional info: I’ve installed tedana in a dedicated conda environment and am running VS code on Windows. Running tedana works (by first tweaking the corresponding “settings.json” file), but somehow debugging is not that straightforward.
Hello! Unfortunately, the developers don’t really use a debugger such as pdb when running tedana. We do have a verbose mode on the command line and in the API, but it doesn’t allow for any interactivity. Could you tell us more about your use case for debug mode?
Thanks for getting back to me.
The main issue I’ve encountered is that either no or very few BOLD-like components are detected while working on a (slice scan time- and motion-corrected) 7T dataset. Some of the rejected components do look rather BOLD-like though, so I wanted to go through the code step by step to find out what exactly is going on (mainly during the component classification).
Based on other posts I’ve already tried 1) setting a different PCA component selection method, and 2) using a different seed for the ICA, but to no avail. What’s more, if I remember correctly, a larger set of components was accepted for this dataset when running tedana v0.0.8 (which I guess cannot be fully attributed to the higher number of retained components due to the now abandoned ‘MLE’ PCA approach).
In short, it would be great to be able to go through the code to better understand what happens ‘under the hood’ when processing a particular dataset.
Thanks again for thinking along!
Hi @exumbra, I’ll jump in on this - regarding 1), changing the pca selection method - which ones did you try? Just trying to get a feel for what you’ve explored here. In addition, any other details about the sequence might be useful - such as the TEs.
The rejected components should also have a reason for their rejection, see here for the look up table - it could be valuable to know if a consistent metric is the reason for rejection.
What @dowdlelt mentions re PCA is an important point. There definitely is a difference in the number of components returned depending on the PCA approach (even among the 3 options in maPCA).
Re understanding what happens under the hood: you could clone the repository and add breakpoints in strategic points in the code you’re interested in.
@dowdlelt and @e.urunuela - thanks for your input!
Regarding the PCA selection methods:
There seemingly is not a lot of dimensionality reduction going on for any of the approaches I tried: 168 (mdl), 171 (kic) and 171 (aic) components are retained for a 172 volumes long (task based) run.
In terms of TEs, we now use TE1/TE2/TE3 = 9.36/23.76/38.16 ms.
The rejected components frequently show ‘rho>kappa’ (i.e., I002), but only with rather slim margins.
I admittedly don’t have experience with repository cloning yet, but will definitely look into this.
Is there any way you could share the data for us to take a look?
I’ll have to check a few things, but in principle that should be doable. Would it be OK for me to contact you by mail to share a link in the upcoming days?
Would actually be great if you could have a look!
Those TEs seem very reasonable for 7T, and cover a nice range.
It may be worth trying the ‘kundu’ method for --tedpca. More details can be found here. I’ve just noticed that this method, along with ‘kundu-stabilize’ isn’t in the command line help but that could be worth exploring. It may lead to a larger difference than what is seen with the three maPCA options, though I am somewhat surprised those three options gave rise to such similar numbers of comps.
@dowdlelt - thanks for the additional info!
Trying the additional pca options certainly led to some interesting results:
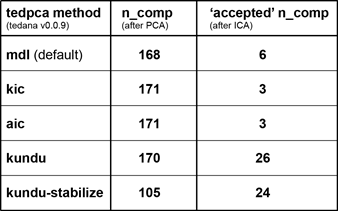
At first sight, the kundu(-stabilize) results appear much closer to the desired outcome (the previously missed BOLD-like components are now accepted and e.g. the range of kappa values is closer to what has been reported in other papers).
It would be interesting to know where these discrepancies originate from, so if it’s OK with you I’ll proceed with making this dataset available to e.g. @jbteves (I guess it’s straightforward to find an email address to share a download-link)?
Thanks again for sharing your thoughts!
Those results are definitely shocking. I would expect MDL to provide the most aggressive dimensionality reduction.
I’d love to see why all methods except kundu-stabilize perform this badly.
You can find any of our email addresses on GitHub (and maybe on our profiles here?).
Thanks for your willingness to have a closer look at this.
I’ve just sent out a mail to the both of you (@jbteves and @e.urunuela) to share this dataset.
Please let me know if you need any additional info or if there’s anything else I could do to help out.
Thanks again and I’m looking forward to hear what you think!
1 Like
Hi @exumbra, I took a look at your data and it’s a bit odd. Your sagittal slices are oriented strangely, and your Nifti header has a Q-form with different handedness from your S-form. How did you do motion correction and slice timing correction? I’m worried that at some point in the process something mangled your data, which could be contributing to this issue.
Thanks for having a look.
The preprocessing was done in BrainVoyager and the data were subsequently exported to Nifti via matlab - admittedly in a rather quick and dirty fashion. I can see whether this could be optimized (apologies if this led to unforeseen issues). However, the very same files were used to try out the different tedpca options mentioned above, so I guess it would still be interesting to see why they diverge so much.
Do you have the un-preprocessed data? I’d like to try and run motion correction on it with AFNI and see if I get a different result, hopefully helping us track down the issue.
I’ve uploaded the unprocessed dicoms and sent you the link.
Thanks again - please let me know if I can help out in any way.
Hi @exumbra. There’s quite a few differences between the data that I get from running a dicom to nifti conversion and then preprocessing with AFNI vs. using your nifti files. The matrix size of the image is different (I get 96x96x81 vs. your 96x96x66), the orientations are very different (which should be irrelevant but indicates some differences in header treatment), and most importantly it seems that despite the same voxel dimensions and the fact that your image seems to be a subset of the one that I got, there are many more nonzero voxels in my image than yours. This is making it a little tricky to do a one-to-one comparison. However, I do believe it’s contributing to the PCA step because you get far fewer samples to conduct it. I’ll continue investigating differences but I wanted to give you a quick update.
Thank you @jbteves for the in-depth look you’re taking.
It could be the case that having many zero values made tedpca fail when performing the subsampling step and calculating the entropy rate (which is used to find the optimal value).
It would be interesting to see how different the two datasets are. Could you share your preprocessed data too @jbteves ?
Thanks @jbteves!
I guess the image matrix size difference is the easiest to explain: our dataset consists of 66 slices, but including the entire dcm mosaic (9x9) would sum up to 81 slices. However, since this would leave you with 15 empty slices at the top, I’m a bit puzzled by your finding that you actually have more nonzero voxels. I’ll try to figure out whether there’s a considerable decrease in nonzero voxels across the different steps within our preprocessing pipeline (in comparison to the original dcm content), but so far I honestly wouldn’t know why that would be the case…
Hm. What are the steps in the pipeline you used?
