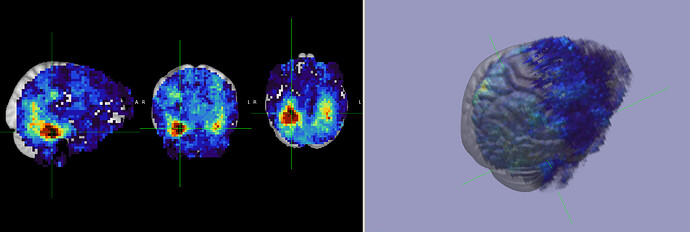
I’m working through the searchlight tutorial.
My latest challenge is shown below. The results of the searchlight are out of registration with the MNI152_T1_1mm_brain standard as shown in FSL.
I would appreciate any insight as to how to correct the registration.
Also, just a sanity check of the results would be very helpful.
Thanks!
hds = get_haxby2001_data_alternative(roi='brain')
hclf = kNN(k=1, dfx=one_minus_correlation, voting='majority')
hcv = CrossValidation(hclf, HalfPartitioner())
hsl = sphere_searchlight(hcv, radius=3, postproc=mean_sample())
hres = sl(hds)
sphere_errors = hres.samples[0]
hres_mean = np.mean(hres)
hres_std = np.std(hres)
chance_level = 1.0 - (1.0 / len(hds.uniquetargets))
frac_lower = np.round(np.mean(sphere_errors < chance_level - 2 * res_std), 3)
map2nifti(hds, 1.0 - sphere_errors).to_filename('hsl.nii.gz')
# Trying reverse mapping to see if registration is correct (no)
revtest = np.arange(100, 100 + hds.nfeatures)
rmapped = hds.a.mapper.reverse1(revtest)
rmapped.shape
#'imghdr' in ds.a
hnimg = map2nifti(hds, revtest)
hnimg = map2nifti(hds, 1.0 - sphere_errors)
hnimg.to_filename('hrsl.nii.gz')