I’m running HALFPipe on an HPC cluster using singularity. The output looks good except for seeing a problem with the EPI image registration to MNI space. There seems to be a registration issue with all of the scans I’m running on multiple occasions (all from the same study but run separately for different scanners), for both resting-state and task-based analyses. This has also been noticed by another researcher using some of the same participants but running HALFpipe separately.
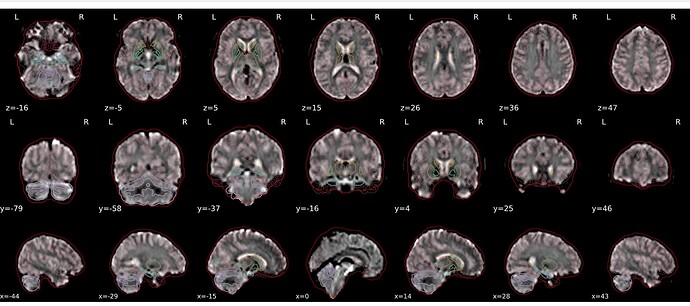
On quality assessment, the brains seem to have “shrunk” where the outline on the EPI SN leaves quite a significant gap between the MNI space template (red outline) and the image.
It was suggested that I enable “recon_all” in the spec.json file, in order to enable Free Surfer as that usually gives better segmentations, which helps with registrations. Unfortunately this did not resolve the issue and the output was the same.
I’m running HALFpipe (version 1.2.2) on a HPC system using singularity. There were no notable error messages or complaints that I could see.
Thought I’d post this here in case someone might have had a similar issue or has any suggestions ![]()