Hello, I’m currently using Nilearn for the analysis of a within-subject fMRI study with a 2x5x2 design. I’ve encountered some issues during the second-level analysis and would greatly appreciate the insights from experts. ![]()
![]()
![]()
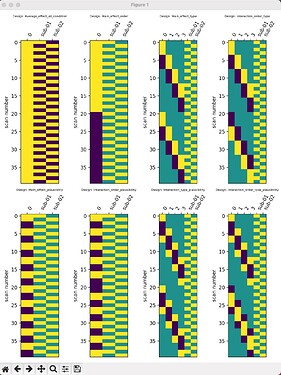
(1) In the first-level analysis, I extracted beta maps for each condition, resulting in 20 beta maps for each subject. The condition arrangement is as follows: A1B1C1, A1B1C2, A1B2C1, A1B2C2, A1B3C1, A1B3C2, A1B4C1, A1B4C2, A1B5C1, A1B5C2, A2B1C1, A2B1C2, A2B2C1, A2B2C2, A2B3C1, A2B3C2, A2B4C1, A2B4C2, A2B5C1, A2B5C2, where A represents the order, B represents the type, and C represents the plausibility. In the second-level analysis, I would like to obtain the main effects of A, B, and C, as well as their interactions. My current design matrix for the second-level analysis, which I generated based on the results from SPM’s function “spm_make_contrasts,” is shown in the figure below.
The contrast matrix was generated using the following code (based on https://en.wikibooks.org/wiki/SPM/Group_Analysis):
all_contrast_matrix = {
'Average_effect_all_condition': [1]+[0]*n_subjects,
'Main_effect_order': [1]+[0]*n_subjects,
'Main_effect_type': [[1,0,0,0]+[0]*n_subjects,[0,1,0,0]+[0]*n_subjects,[0,0,1,0]+[0]*n_subjects,[0,0,0,1]+[0]*n_subjects],
'Interaction_order_type':[[1,0,0,0]+[0]*n_subjects,[0,1,0,0]+[0]*n_subjects,[0,0,1,0]+[0]*n_subjects,[0,0,0,1]+[0]*n_subjects],
'Main_effect_plausibility': [1]+[0]*n_subjects,
'Interaction_order_plausibility': [1]+[0]*n_subjects,
'Interaction_type_plausibility': [[1,0,0,0]+[0]*n_subjects,[0,1,0,0]+[0]*n_subjects,[0,0,1,0]+[0]*n_subjects,[0,0,0,1]+[0]*n_subjects],
'Interaction_order_type_plausibility': [[1,0,0,0]+[0]*n_subjects,[0,1,0,0]+[0]*n_subjects,[0,0,1,0]+[0]*n_subjects,[0,0,0,1]+[0]*n_subjects],
}
Could you please confirm whether my design matrix and contrast matrix are correct for achieving my intended results?
(2) Additionally, there’s another within-subject condition D (task with 2 levels). Can I build on my current setup and perform another second-level analysis (effectively a third level) to incorporate condition D? This would allow me to investigate whether the observed effects change, disappear, or remain consistent across the different levels of D.
(3) I’m also unsure about the output_type parameter in nilearn.glm.first_level.FirstLevelModel.compute_contrast. How do ‘z_score’, ‘p_value’, and ‘effect_size’ differ, and why is ‘effect_size’ (i.e., beta) used in the first-level analysis and then fed into the second-level model? Is it possible to use ‘z_score’?
I’m relatively new to fMRI analysis, so please pardon the simplicity and naivety of my questions. I sincerely appreciate your assistance in advance!