Hi experts,
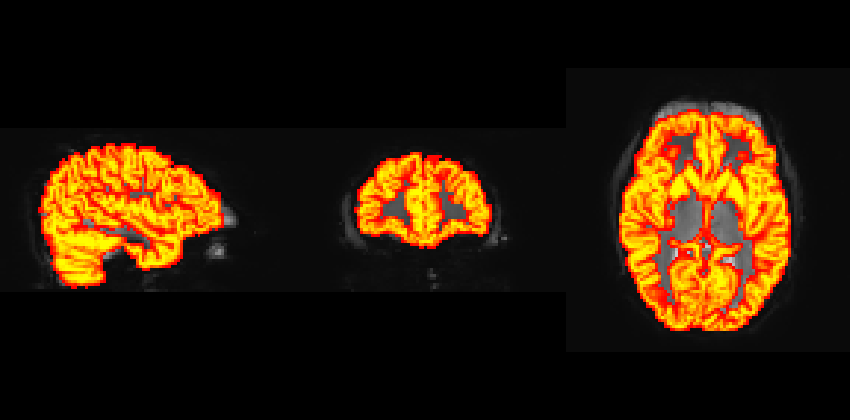
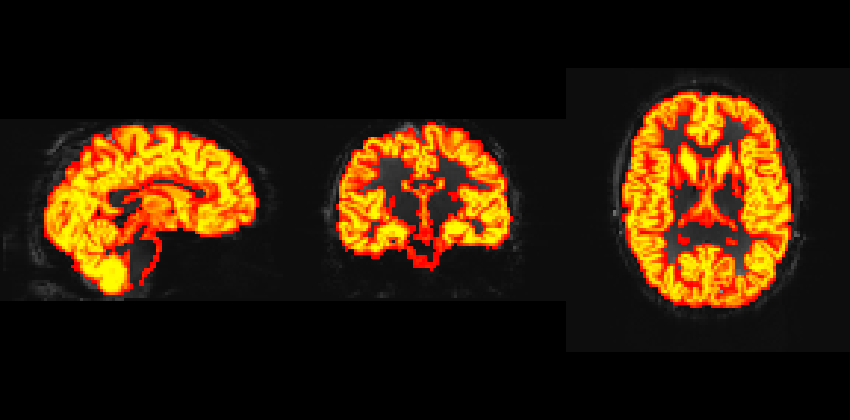
I have registered the GM mask/T1 to EPI space for both Philips and Siemens data. However, I noticed that the GM mask does not overlay exactly on the EPI image from the Philips data, particularly in the frontal areas where distortion corrections were applied. The data was preprocessed using fmriprep.
Below are the overlay images:
GM overlay on EPI from the Philips scanner.
GM overlay on EPI from the Siemens scanner.
Does it seem like the registration was poorly done on the Philips data? though the same registration process worked fine on the Siemens data. Below is the registration process I applied:
#register the T1 image to the EPI using the middle volume as the reference with FLIRT
flirt -in "$brain_mask" -ref "$derivatives_folder/${subject_id}_ses-${session_id}_task-CVR_desc-preproc_reorient_midvol_brain_bold.nii.gz" -out "$derivatives_folder/${subject_id}_ses-${session_id}_T1_to_EPI.nii.gz" -omat "$derivatives_folder/${subject_id}_ses-${session_id}_motion_corrected.mat" -dof 6 || { log "Error in registering T1 to EPI"; exit 1; }
# Apply the transformation matrix to GM probability map
flirt -in "$derivatives_folder/${subject_id}_ses-${session_id}_label-GM_binseg.nii.gz" -ref "$derivatives_folder/${subject_id}_ses-${session_id}_task-CVR_desc-preproc_reorient_midvol_brain_bold.nii.gz" -out "$derivatives_folder/${subject_id}_ses-${session_id}_label-GM_binseg_in_EPI.nii.gz" -applyxfm -init "$derivatives_folder/${subject_id}_ses-${session_id}_motion_corrected.mat" || { log "Error in applying transformation matrix to GM probability map"; exit 1; }
Could you please advise if there is anything wrong with these steps, or suggest ways to improve the registration for the Philips data?
Thank you very much for your help.
Best regards,
Ru