Summary of what happened:
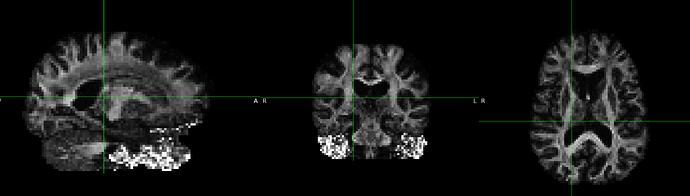
Here is a FA image I tried to align to the standard FA
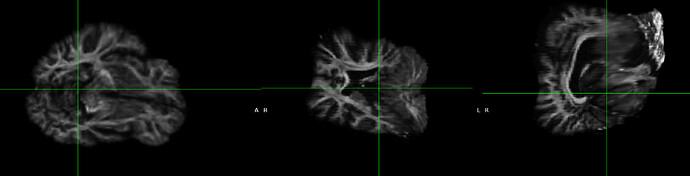
Here is the final results:
Command used (and if a helper script was used, a link to the helper script or the command generated):
First used dcm2niix to do the conversion:
dcm2niix -a y -d n -g y -z y -o
Then process through a standard fdt pipeline;
Copy all FA images to a TBSS folder, then run
tbss_1_preproc
tbss_2_reg -T
tbss_3_postreg -T
People suggested to do a reorientation for the FA image, but I tried that and didn’t work. My images appear to be in the right orientation and fslreorient2std didn’t do anything.
FSL TBSS question… Warping? - Neuro Questions - Neurostars
Version:
fsl 5.0.3