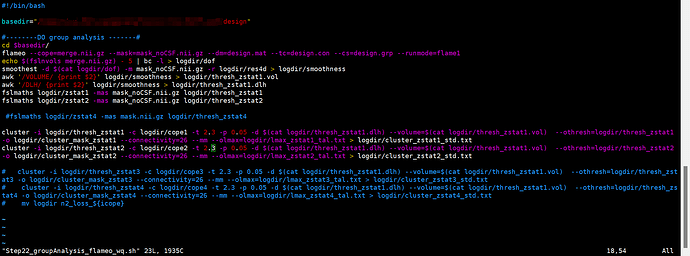
I recently conducted GLM analysis using the indicator (fALFF) calculated from the resting-fmri data of 429 participants to verify the relationship between this indicator and the scale score. The following is the code for running the analysis.
As a normal result, this line of code (smoothest -d $(cat logdir/dof) -m mask_noCSF.nii.gz -r logdir/res4d > logdir/smoothnes) will output a file called smoothness, which contains a DLH value that will be used for subsequent clustering correction, but this value is actually NaN (as shown in the figure). What is the reason for this? And how will this affect the results of cluster correction ?