Dear all,
I have processed multi-echo resting-state fMRI images (three echoes per scan) using fMRIPrep (V.24.1.0) on singularity. To remove dummy volumes and perform further denoising, I applied the following steps on fmriprep outputs:
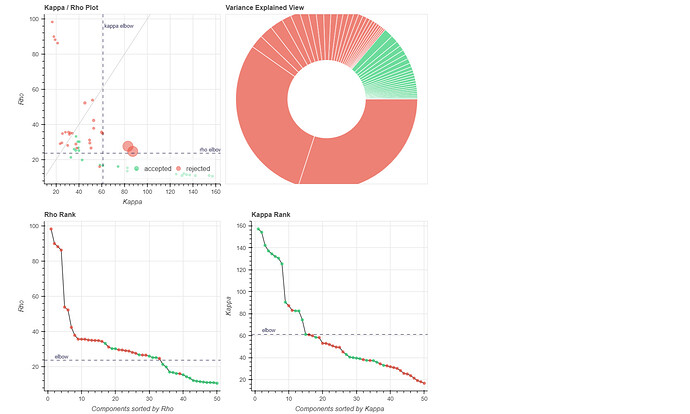
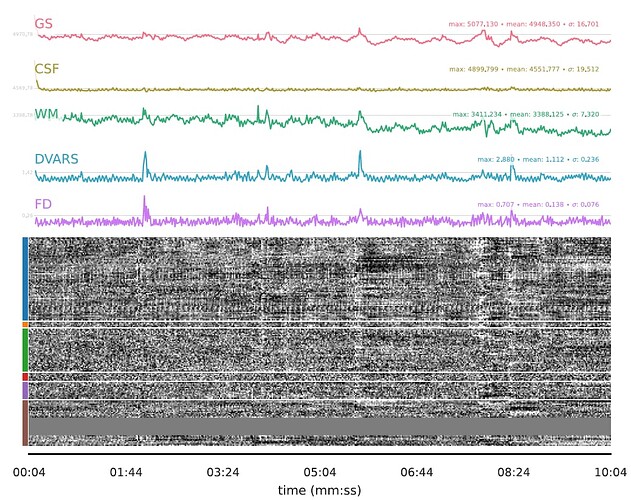
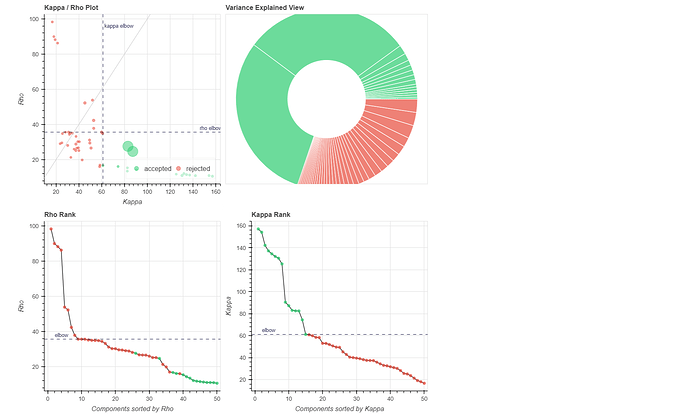
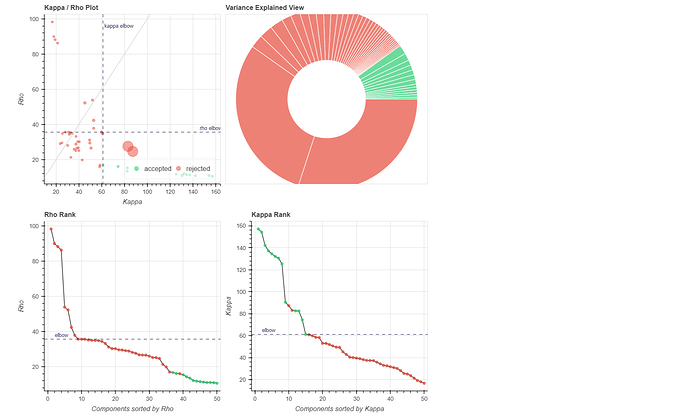
(1) removed the dummy volumes from the echo-wise files, (2) ran tedana (V 24.0.2) on those trimmed files, (3) removed the dummy volumes from the MNI-space optimally combined file from fMRIPrep, and (4) used tedana ICA components to denoise the trimmed MNI-space file as discussed and suggested on a (former topic on Neurostarts). I took the non-aggressive approach with no confounds (just the rejected components) to apply denoising on the data using Nilearn. After denoising, I extracted the timeseries from the Schaefer 7-200 parcellation and computed the corresponding functional connectivity matrices. However, the resulting connectivity matrices exhibit unusually high values, which seem unexpected to me. I’d appreciate any insights on whether such high connectivity values are typical given this type of sequence and denoising approach. If they are expected, are they considered acceptable? If not, what strategies could be used to address this issue?
Attached below are the codes used for these steps, including the application of Tedana on fMRIPrep outputs to obtain the main mixing matrices necessary for further image denoising:
tedana_wave2.txt (3.7 KB)
Code for applying tedana denoising using nilearn:
denoise_nonaggr_MTL0002.txt (2.8 KB)
Code to extract connectivity matrices from the Schaefer parcellations:
connectivity_schaefer200_wave2_MTL0002_FU84.txt (2.1 KB)
Here is the connectivity matrix csv file:
connectivity_nonaggr_schaefer200_MTL0002_FU84.txt (978.4 KB)
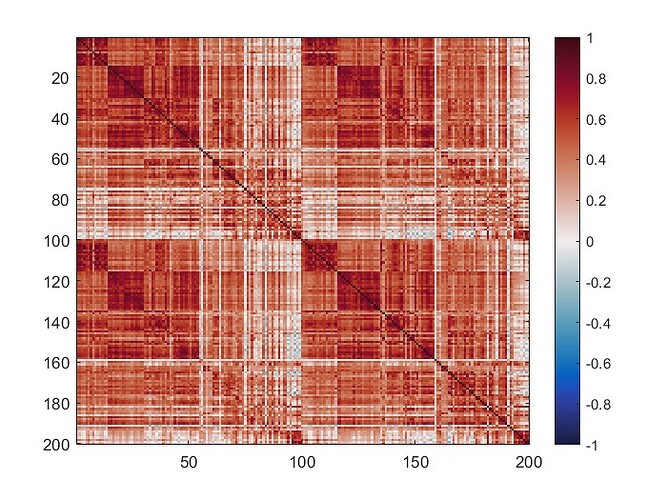
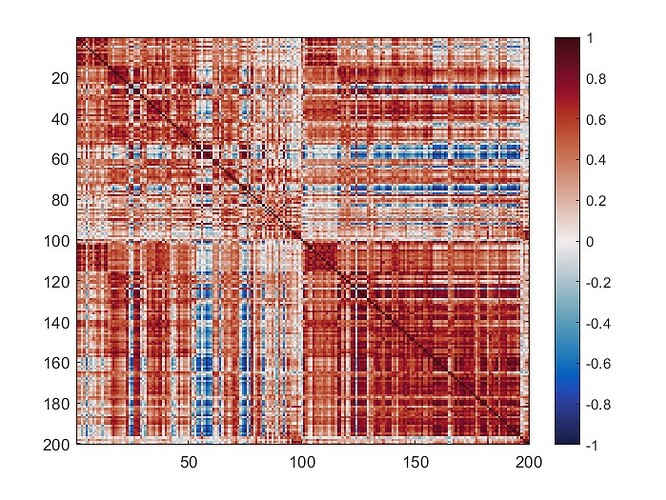
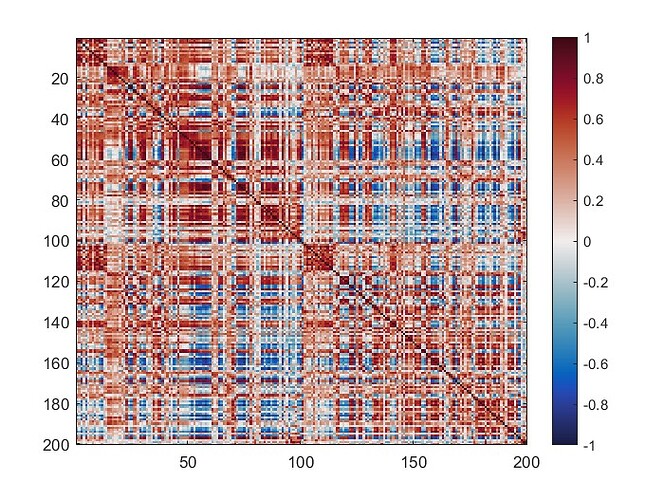
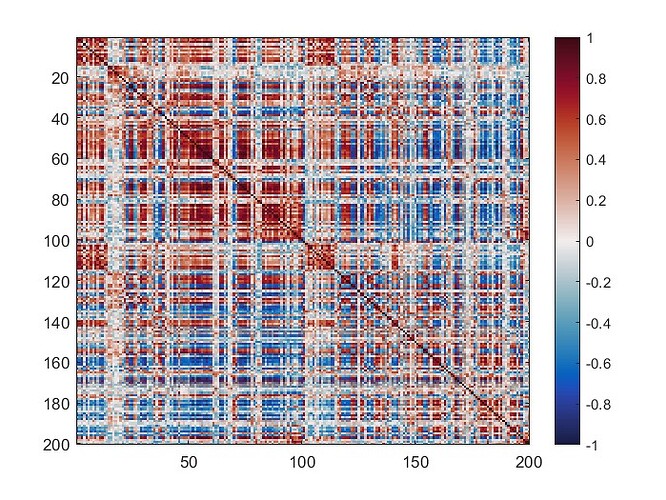
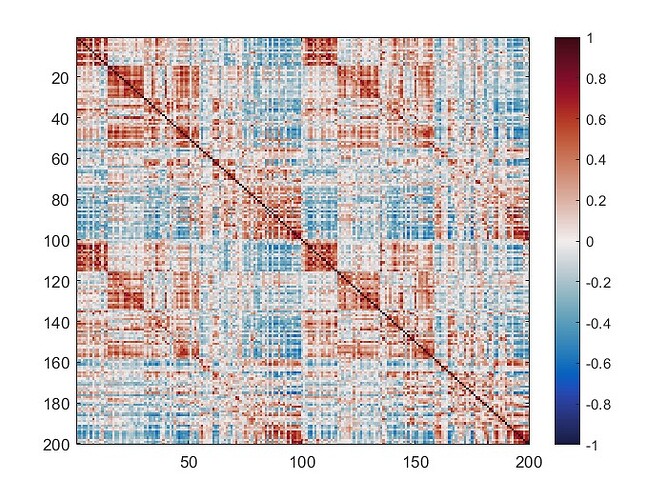
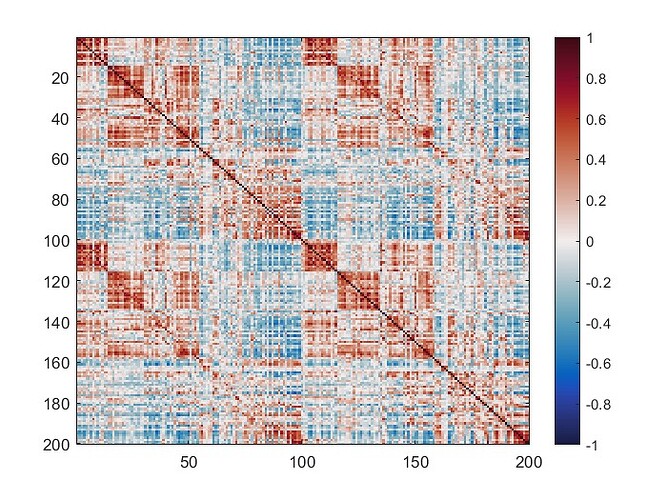
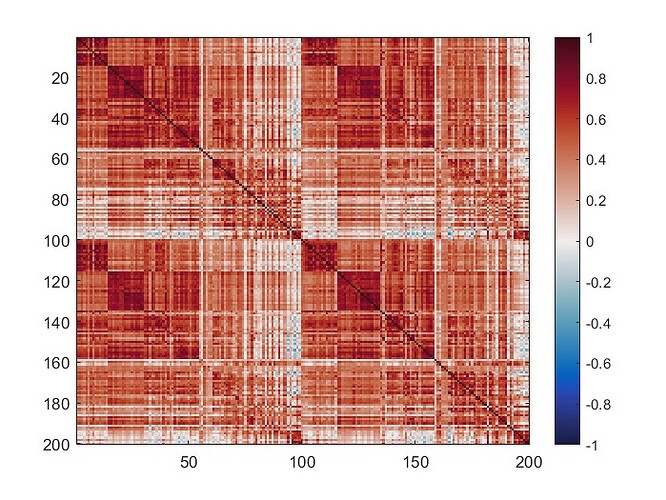
The mean connectivity value of the whole matrix is 0.42, and here is the plotted functional connectivity matrix.
The following image is the plotted connectivity matrix in matlab.
Many thanks in advance,
Ali