Summary of what happened:
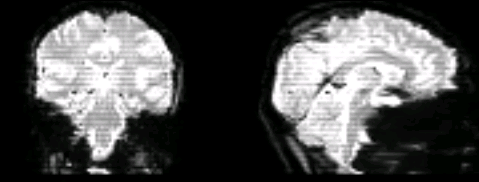
I have fMRI dataset with a striping artefact along the Z axis due to the acquisition sequence.
I am using fMRIPrep 23.1.4 to motion correct and create the registrations to MNI space etc, and FEAT 6.00 to do the stats analysis.
To work around the striping issue, I have done the first level analysis in naitive space (func). For the second level analysis, I now need the registrations to MNI, but these dont seem to have come from fMRIPrep in a form useable by FEAT.
The only transform files I have found to MNI are the sub-001_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5 transformations, which are for the T1 image and in a .h5 format, whereas the FEAT documentation indicates using updatefeatreg, after replacing some or all of: example_func2highres.mat, highres2standard.mat, example_func2highres_warp.nii.gz, highres2standard_warp.nii.gz, example_func2initial_highres.mat or initial_highres2highres.mat
I have output spaces for fMRIPrep as rest and MNI152NLin2009cAsym.
I am aware of the Mumford workaround but my understanding is that they do the first level analysis in standard MNI space - hence the identity transformation between first and second level analyses - whereas I will need to add the func → MNI transformations between levels 1 and 2 of FEAT (steps 5-6 in the workaround).
Can I use fMRIPrep to make the func → MNI transformations?
Is there another way I should be approaching this?
Thanks in advance
Screenshots / relevant information:
The scans look like this: