Yes, thank you for offering the suggestion of adding the option -fout fieldmap in topup! I think that would be very useful. Please note however that it would be most useful if the acqparams.txt file is filled with the correct "TotalReadoutTime" value of the distorted EPI image used for topup with synbold-disco, this way the “fieldmap” (in Hz) created could be used with tool such as fmriprep that look at the accurate "TotalReadoutTime" of the distorted EPI scans to apply the deformation field for SDC.
Hi @jsein,
I was wondering if you could provide a little bit more guidance about the files you need to replace inside fMRIprep for this approach to work. I have two magnitude images + the phase difference but I’m not particularly happy with the result and I am interested in trying this approach. Thanks in advance.
Best regards,
Manuel
This is indeed interesting! We’ve only started validation on the final distortion-corrected images and not the field maps themselves - but I also do not like the way that field map looks. Let me reproduce this and debug. We are also using the b02b0.cnf so it seems the only differences would be the three options (subsamp, miter, lambda) that we are over-riding in the topup call. Until then, we will recontainerize and add the field map as output, and also enable a user input for acqparams.txt so that the map units are meaningful.
Thank you,
Kurt
Hi @mblesac ,
Which fmriprep version are you working with? As you must have noted (and perhaps wrote about it already) there are several posts covering this issue with SDC not working well in FMRIPREP, especially for the “Direct Fieldmap” approach (Two magnitude and one fiedlmap images). Some corrections were made for version 23.0.0, perhaps you could try it?
As it was rightfully stated by @schillkg , synbold-disco was designed for cases where no fieldmap were acquired: normally if all goes well, SDC should work at least as well as synbold-disco for bold images for which proper fieldmap were acquired.
Anyway, if you want to give a try to the synbold-disco/fmriprep method for you dataset, for the method that worked correctly in my test, here are the steps I followed:
1/ run synbold-disco for your distorted bold serie. You must rename your bold image as: BOLD_d.nii.gz and your T1w image as: T1.nii.gz and place those in the INPUTS directory. Then run synbold-disco. In my case I used singularity with the following command:
singularity run -e \
-B /scratch/jsein/BIDS/$study/sourcedata/synbold/sub-${sub}/INPUTS:/INPUTS \
-B /scratch/jsein/BIDS/$study/derivatives/synbold/sub-${sub}:/OUTPUTS \
-B /scratch/jsein/BIDS/freesurfer/license.txt:/opt/freesurfer/license.txt \
/scratch/jsein/my_images/synbold-disco-v1.3.simg --stripped --motion_corrected
2/ For the moment, synbold-disco doesn’t call topup with the --fout option to output the fieldmap in Hz. So I proceeded by calling topup once more on the OUTPUTS of synbold-disco, through singularity again, using my edited acqparamsJS.txt file:
cat acqparamsJS.txt
0 1 0 real_total_readout_time_of_bold # my bold image are with PE direction in P->A
0 1 0 0
singularity exec -e -B /scratch/jsein/BIDS/$study \
/scratch/jsein/my_images/fmriprep-23.0.0.simg \
topup --imain=/scratch/jsein/BIDS/$study/derivatives/synbold/sub-${sub}/BOLD_all.nii.gz \
--datain=/scratch/jsein/BIDS/$study/derivatives/synbold/sub-${sub}/acqparamsJS.txt \
--config=b02b0.cnf --subsamp=1,1,1,1,1,1,1,1,1 --miter=10,10,10,10,10,20,20,30,30 \
--lambda=0.00033,0.000067,0.0000067,0.000001,0.00000033,0.000000033,0.0000000033,0.000000000033,0.00000000000067 --scale=0 \
--out=/scratch/jsein/BIDS/$study/derivatives/synbold/sub-${sub}/my_topup_results \
--fout=/scratch/jsein/BIDS/$study/derivatives/synbold/sub-${sub}/my_field \
--iout=/scratch/jsein/BIDS/$study/derivatives/synbold/sub-${sub}/my_unwarped_images \
--jacout=jac --rbmout=xfm --dfout=warpfield -v
3/ Rename the files needed for SDC within FMRIPREP according the “direct fieldmap” method:
-
my_field.nii.gz(created in step 2/) →fmap/sub-SUB_ses-SES_fieldmap.nii.gz - copy the JSON file from your func image into
fmap/sub-SUB_ses-SES_fieldmap.jsonand add the following tags:
"Units": "Hz",
"IntendedFor": [
"ses-SES/func/sub-SUB_ses-SES_task-TASK_bold.nii.gz"
]
(you could also use the B0FieldIdentifier / B0fieldSource strategy).
-
BOLD_s_3D.nii.gz→fmap/sub-SUB_ses-SES_magnitude.nii.gz
Now you are all set to run FMRIPREP using the fieldmap estimated the synbold-disco!
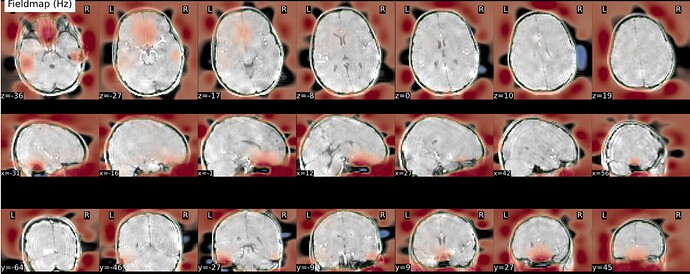
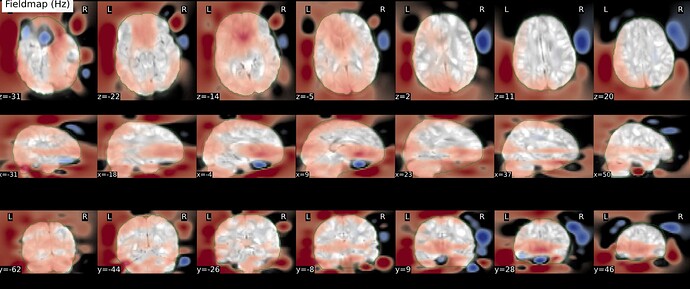
As stated above, the fieldmap calculated by synbold-disco has a strange appearance but the correction looked correct (at least for the one subject I tried so far).
I can’t guaranty it will work for you but it would be interesting to see if you get good results for your dataset with this method.
Hi @jsein,
Thanks for your help. I followed your instructions and I finally have the results.
This is 5 years old children’s data, so I executed the synbold-disco slightly differently, I add the flag --no_topup and removed the flags --stripped and --motion_corrected.
My acqparams file is also different as the phase encoding is j- so I replaced the 1 for -1.
The fMRIprep call I used in both cases is:
singularity run -C -B /EBC:/EBC,$PWD:/opt/templateflow /EBC/home/mblesa/fmriprep-23.0.0.simg $PWD/BIDs_data $PWD/derivatives/sub-${NAME}_fmriprep participant --participant-label ${NAME} -w $PWD --nthreads 16 --fd-spike-threshold 2 --dvars-spike-threshold 3 --omp-nthreads 16 --fs-license-file /EBC/local/infantFS/freesurfer/license.txt --output-space MNI152NLin2009cAsym:res-2 T1w --stop-on-first-crash
Here the results:
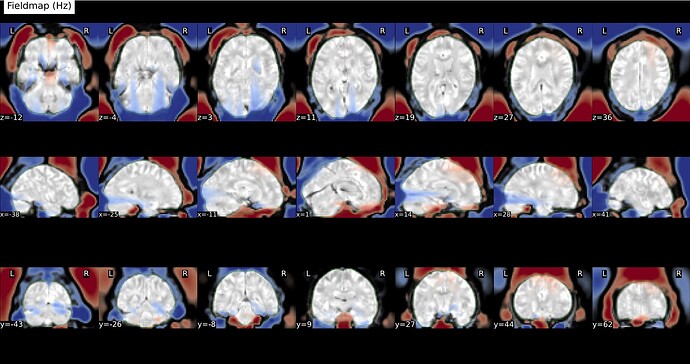
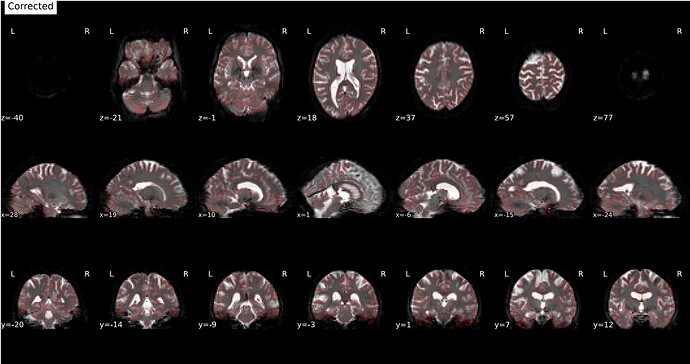
Using the two magnitude images + phase difference:
Fieldmap:
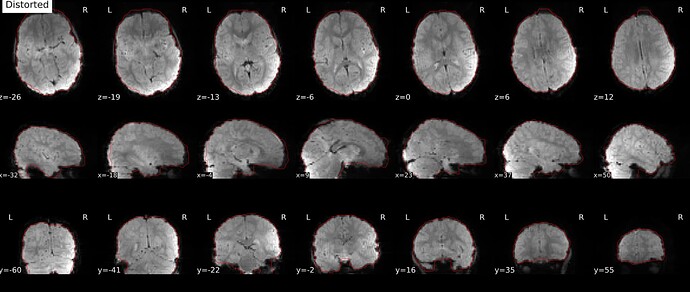
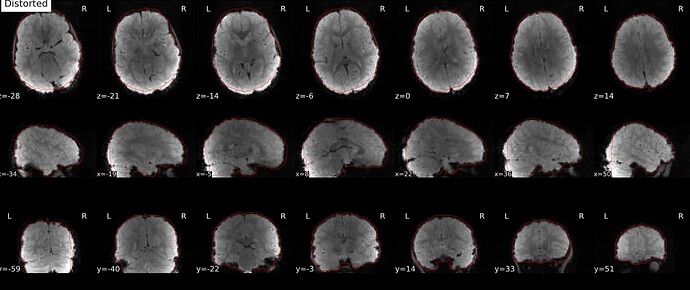
Distorted image:
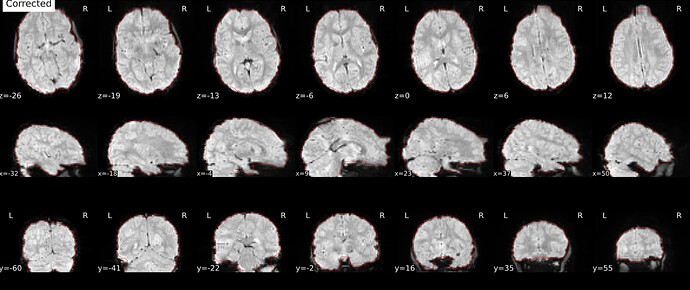
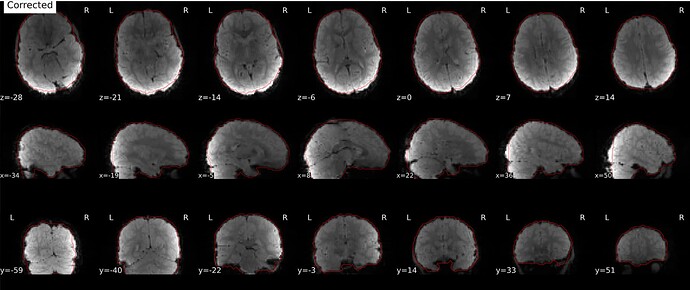
Corrected image:
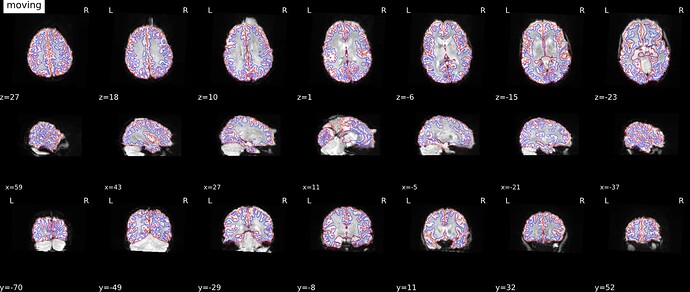
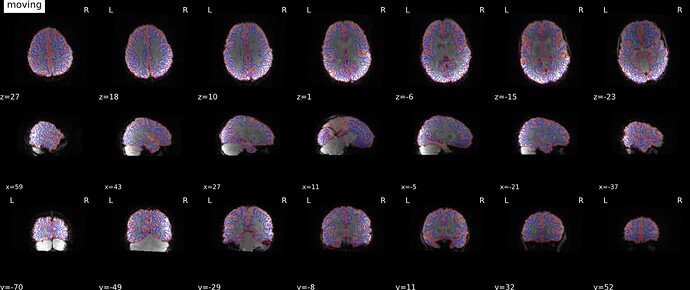
Aligned:
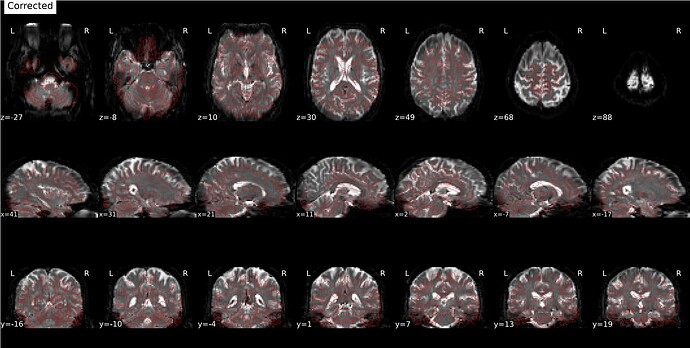
Using the fieldmap generated from Synbold-disco:
Fieldmap:
Distorted image:
Corrected image:
Aligned:
I think the Synbold-disco looks much better. What do you think?
If I want to run this process for different fMRI task (acquisitions) should I run Synbold-disco for each one? or use the same fieldmap for all of them? (tagging @schillkg here)
One small last question, why in some cases there are intensity differences between the corrected and the distorted images?
Thanks in advance!
Best regards,
Manuel
Hi all - quick update - the container is currently rebuilding to (1) output the field map directly and also to (2) enable a user-defined acqparams file (actually just a TotalReadoutTime variable) so that the field map has meaningful units. We may actually kick off another docker build to also allow a user defined .cnf file if one chooses to modify some of the topup parameters. We want to run a quick couple of tests but will hopefully upload this by the weekend or early next week. This will let me run some tests to see why our field map looked a bit unconventional (higher frequency + streaks?).
@mblesac - One could argue that you should do whatever is most conventional for a traditional analysis as if you did have field maps of reverse PE available. In most cases (just browsing some datasets from openneuro) it seems only a single field map or single reverse PE pair is acquired prior to all FMRI runs. So I believe it would be common to use the field map (or field map estimated from the rPE pair) for all subsequent scans, assuming contrast and bandwidth stay the same. However, if you have reason to believe that the field map may change over a period of time across runs (movement? hardware drift?) then I see no reason not to estimate the field for each run separately - although I have no expertise in FMRI and preprocessing!
One important note: if you do have reverse PE or field maps available we still suggest using these! It’s not that we don’t believe in our algorithm, we are just trying to make it clear that we aren’t claiming we are “better” in any way. In fact, we are just using tricks to estimate a field map through image processing rather than image acquisition (and use topup itself in doing so)!
Could you explain the intensity differences you are seeing? I see bias field present in both distorted and corrected images (which make sense because we don’t correct for this at this point). Applying the topup field map will also correct small signal stretch/pile-up, although this is much less noticeable in gradient echo images than in the diffusion spin echo I’m used to working with! Finally, we have the --scale option in topup set to 1, where topup will rescale each image to the individual mean. This could also cause an intensity difference, although it would be a single multiplicative factor for the entire volume.
@mblesac and @jsein are the field map visualizations and alignment with t1 + edges a QA output of fmriprep? These are outstanding QA visualizations and have convinced me to make sure any future updates are easily integrated with other packages!
@mblesac : I agree to what @schillkg is saying about the number of fieldmaps to use for each subject. One is generally fine, you may want to use more in cases where the subject changes his head position between runs of if he moves in and out of the scanner during one session.
Yes indeed those nice visualization figures are provided in the HTML report after a fmriprep execution, there are very useful to quickly assess how the fmriprep workflow went.
More details here:
https://fmriprep.org/en/stable/outputs.html#visual-reports
Thank you @schillkg for providing and improving the Sync-disco ans Synb0-disco tools!
** Sorry this will be split into 3 posts because new users can only embed 1 image file in a post**
Hi all,
I have some insight into the field maps. Using a few datasets from openneuro (results happen to be from ds004158 OpenNeuro, but seems to generalize to a few others: ds004215, ds003507) - I’ve run topup using our synbold parameters (which overwrite --subsamp, --miter, --lambda in the default cnf file), as well as topup using the standard b02b0.cnf, and also topup using the traditional (our silver standard) forward and reverse PE images.
To recap:
-
topup -v --imain=BOLD_all.nii.gz --datain=acqparams.txt --config=b02b0.cnf --iout=BOLD_all_topup --out=topup_results --fout=topup_field --subsamp=1,1,1,1,1,1,1,1,1 --miter=10,10,10,10,10,20,20,30,30 --lambda=0.00033,0.000067,0.0000067,0.000001,0.00000033,0.000000033,0.0000000033,0.000000000033,0.00000000000067 --scale=0
-
topup -v --imain=BOLD_all.nii.gz --datain=acqparams.txt --config=b02b0.cnf --iout=BOLD_all_topup --out=topup_results --fout=topup_field
-
topup -v --imain=BOLD_all.nii.gz --datain=acqparams.txt --config=b02b0.cnf --iout=BOLD_all_topup --out=topup_results --fout=topup_field
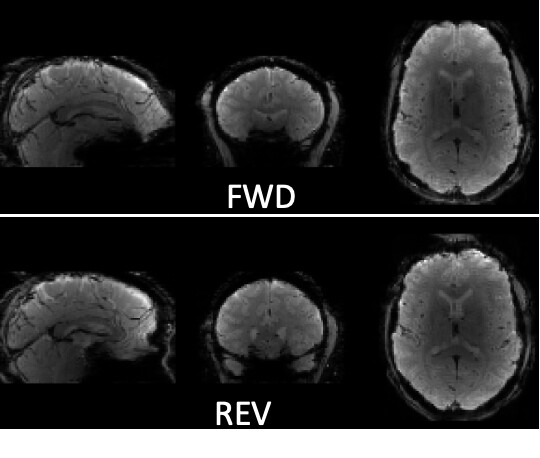
Here #1 and #2 the BOLD_all is composed of FWD-distorted+synthetic, whereas for #3 it is FWD-distorted+REV-distorted.
Result #1. With default contrast limits on fsleyes (which I think set limits to image min and max), little qualitative differences:
Results #2: adjusting contrast, I see this streaking in all the field maps, even with topup run the traditional way! I did not expect this, however I have never fully inspected the field maps (nor the stored coefficients). However, the original implementation with less regularization and no subsampling was ‘streakier’.
Result #3: the final application of these maps is remarkably similar regardless:
We did find one interesting result that the non-subsampled topup (the first algorithm) uniformly shifted the voxels in the anterior direction forward by 1 unit, but otherwise resulted in qualitatively similar distortion correction.
So it seems that the distortion correction is similar despite differences in appearance of the field map…and optimistically, ‘similar’ to as if a full reverse PE scan was available! We are going to do a few things: (1) we will always output the field map (–fout) in addition to the coefficients (2) we will default to b02b0_1.cnf when data matrix is odd and b02b0_2.cnf when data matrix is even (we will remove our parameter tweaks, that we had previously optimized for the spin echo diffusion data). (3) we will make a TotalReadoutTime input so that the field map has meaningful units. And (4) one thing not previously discussed: we are going to enable the ability to turn off the smoothing we do on the input images.
Finally - we hope to fully validate this soon with quantitative comparisons to T1, in a manner similar to another approach for fieldmapless correction (Using synthetic MR images for distortion correction - ScienceDirect which uses nonlinear registration, as opposed to our approach of using the physics-based estimation within topup to derive a field map).
Happy to further debug (and of course improve) this approach!
Thank you,
Kurt
Hi all,
We wanted to post a quick update on this thread. The synthetic field map algorithm designed specifically for gradient echo fMRI is now published (https://t.co/ZtBGxKXBd1) and available (https://github.com/MASILab/SynBOLD-DisCo). We have made it possible to output the field map directly, and also allow user-defined TotalReadoutTime.
Have you all had successes failures with integrating into fmriprep? If successful, we would love to put a step-by-step guide on our github page (or if anyone has connections we’d be happy to try to integrate directly into fmriprep itself).
Thank you again,
MASI lab team
Hi @Bennett1, and welcome to neurostars!
I believe a basic step-by-step is something like:
Using RAW data:
- Run SynBOLD-DisCo without topup (
--no_topup) - Take the synthesized undistorted BOLD-contrast image (
BOLD_s_3D.nii.gz), place in thefmapfolder, give it a name such assub-xx_ses-xx_acq-synbold_dir-PA/AP_epi.nii.gz, where thedir-<>label is the opposite of what your original BOLD is. - Make your own JSON. Just needs a few fields: (1)
TotalReadoutTime- make it whatever you define at runtime or the default (1 second), (2)PhaseEncodingDirection- make it the opposite as your original image (so add or remove the minus sign), (3) Add theIntendedForand/orB0FieldSource/Identifierfields to link the fieldmap to your BOLD image. The JSON should be calledsub-xx_ses-xx_acq-synbold_dir-PA/AP_epi.json. - Run fMRIPrep as normal (of course, do not
--ignore fieldmapsor--use-syn-sdc, otherwise the fmaps may be not used)
Best,
Steven
EDIT: While this probably works fine with just 1 T1 image, I now wonder if it would be better to use the preprocessed T1 template in the case of multiple T1s. That would make the workflow a little more complicated, since it would involve something like running sMRIPrep (of fMRIPrep with --anat-only) first, then making the synthesized image, and then doing the rest of fMRIPrep (passing in the preprocessed anatomical derivatives with the --anat-derivatives flag). I might give this a shot since I have a 7T dataset with multiple T1 images.
Hi all,
Thanks to all the contributors to this interesting and useful thread! I have tried fmriprep23.# for a couple of fMRI runs either using its fieldmap-less or with fieldmap susceptibility distortion correction on a couple of datasets, however they all made more distortions (discussed in a thread here). Thus, I am now trying SynBOLD to see whether I can get a detour for that SDC part of fmriprep. Also, I am currently using an old dataset with no fieldmap data available.
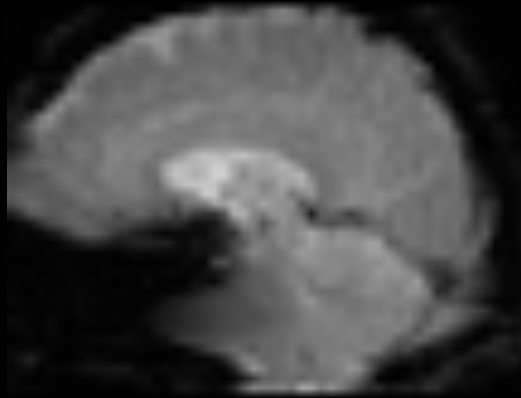
So far, results from the SynBOLD (BOLD_u.nii.gz) are amazingly good (by means of distortion and matching with T1), however, while trying to utilize it into fMRIprep I encountered some issues:
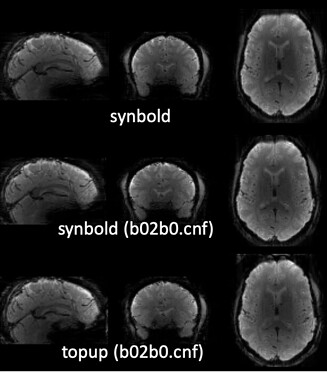
-
Using the method by @Steven did not solve the issue. Of note, it can be also because of my new fmriprep version and its buggy SDC. Check the frontal and brainstem regions:
-
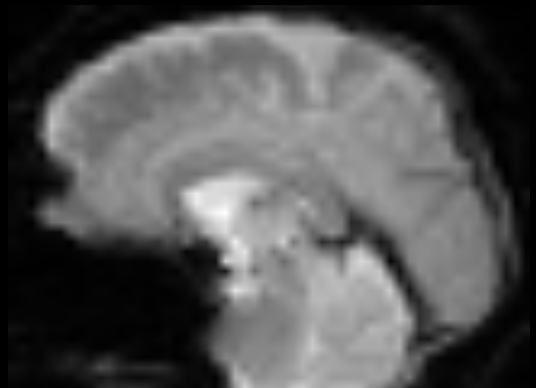
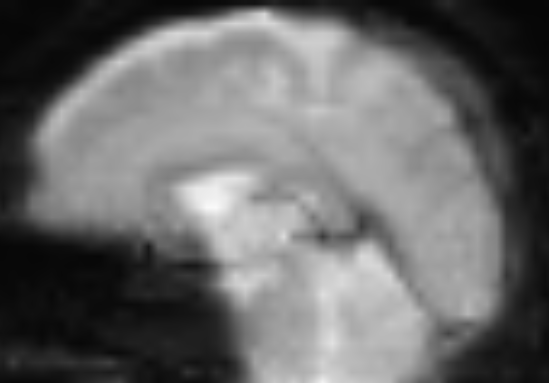
I ran the pipeline suggested @jsein, with slight difference:
– Letting TOPUP be done by SynBOLD
– No --stripped or --motion_correct flags; I am also curious why these flags were used in the first place if the data was not either stripped or motion-corrected.
The results of SDC ran by fmriprep (sub-#_ses-#_task-rest_space-T1w_desc-preproc_bold.nii.gz) are better (top image) but still worse than the BOLD_u.nii.gz generated by SynBOLD (bottom image). Both are in the T1w space for better comparison:
Is there a way to pass the BOLD_u.nii.gz to the fmriprep and do not have the issues mentioned here? I haven’t tried it yet but was thinking of:
- Running SynBOLD with --no_smoothing flag to avoid double-smoothing
- Pass BOLD_u_3D.nii.gz as “sbref” in the fmriprep to be used for mcflirt and motion regressor estimation.
- Use rBOLD.par to de-“motion correct” the BOLD_u.nii.gz. However, I do not know whether it is possible or not.
I also wonder whether it is possible to run fmriprep on BOLD_u.nii.gz and pass the rBOLD.par into the confounds pipeline of fmriprep separately.
Best,
Amir
Hi @Steven!
Sorry for reviving this old topic, but since I ran into issues with this specific approach, I wanted to ask for some clarification.
I tried the approach you suggested, but while the SynBOLD-DisCo results looked good (in terms of correcting the suceptibility distortions), the resulting func files in fmriprep were still distorted. Specifically, in the fmap folder I gave the original AP image (with PhaseEncodingDirection": "j") collected for topup and the synthetic BOLD from SynBOLD-DisCo with dir-PA (with PhaseEncodingDirection": "j-"). Therefor, I wonder:
-
Why use a
TotalReadoutTimeof 1 second? Shouldn’t we give a value as close to 0 as possible, since the synthesized image approximates an acquisition with infinite bandwidth? -
Is there a reason not to run SynBOLD-DisCo with topup and simply give the output fieldmap
topup_results_field.nii.gzwith a json file of the form:
{
"Units": "Hz",
"IntendedFor": "func/sub-01_task-rest_bold.nii.gz",
"B0FieldSource": "topup",
"PhaseEncodingDirection": "j"
}
One problem might be that fmriprep also expects to get a magnitude image along with this fieldmap, if I understand correctly.
Perhaps @schillkg is the person to ask?
Any help would be greatly appreciated!
Thanks,
Roey
P.S.
It just occured to me that @jsein gave the solution above, so I will give it a try!
Hi @roeysc - I can’t speak to fmriprep particulars, but can hopefully give some overall insight.
The total readout time for the synthesized image should indeed be 0 (infinite bandwidth).
This next statement has some caveats: The total readout time for the acquired (distorted) image doesn’t matter, and we typically default to something like 0.05. If you were to use topup to estimate the field, then applytopup to apply this field to the unwrapping process, as long as you give applytopup the acquisition parameters (i.e., the total readout time you gave topup) it will correctly scale the field.
However, if the units/magnitude of the field map matters for subsequent processing, then the total readout time matters. I’m not sure how fmriprep chooses to estimate and apply the field, so this value possibly does matter.
We also also recommend using reverse PE or field maps when those are available, and only using the synthesis method when these are unavailable. Hope that helps answer some of the questions.
Hi,
Apologies for reviving this old topic. I tried the approach mentioned above:
The SynBOLD outcome (BOLD_s_3d.nii.gz) seems quite nice:
But the results of fMRIprep (v 25.0.0) are not promising:
Note the extended blue areas into the brain:
Note the shady areas in the frontal, occipital, and brainstem regions. It was more severe in some other subjects.
And for another subject:
Here is the fMRIprep command in its report, ran by Singularity:
/opt/conda/envs/fmriprep/bin/fmriprep /input/ /derivatives/fmriprep_25.0.0_nosdc --skip_bids_validation --fs-license-file /toolboxes/license.txt participant --participant-label sub-01 -t rest --output-spaces MNI152NLin6Asym:res-2 MNI152NLin2009cAsym -w /work/ --stop-on-first-crash --nthreads 8
I have not tried fMRIprep’s “syn” pipeline per se, and I have no experience with which approach, SynBOLD or fMRIprep’s Syn, results in better SDC. Any guide/help is appreciated.
Best,
Amir
I’m a bit confused here, shouldn’t this step
- Take the synthesized undistorted BOLD-contrast image (
BOLD_s_3D.nii.gz), place in thefmapfolder, give it a name such assub-xx_ses-xx_acq-synbold_dir-PA/AP_epi.nii.gz, where thedir-<>label is the opposite of what your original BOLD is.- Make your own JSON. Just needs a few fields: (1)
TotalReadoutTime- make it whatever you define at runtime or the default (1 second), (2)PhaseEncodingDirection- make it the opposite as your original image (so add or remove the minus sign), (3) Add theIntendedForand/orB0FieldSource/Identifierfields to link the fieldmap to your BOLD image. The JSON should be calledsub-xx_ses-xx_acq-synbold_dir-PA/AP_epi.json.
use a TotalReadOutTime of close to 0? The idea is the synthesized image is close to undistorted, so it should have very small total readout time. For DWI, I have done
‘TotalReadoutTime’: 0.0000001
‘EffectiveEchoSpacing’: 0.0
Hi all,
I am trying to do the same - use the fieldmaps generated by SynBOLD-disco with fMRIprep. I trialled renaming the top-up outputs and putting them into the sub-XX/fmap/ folder.
I’m a bit confused as to what goes where and what info goes into the json.
The topup_results_field.nii.gz is the same dimensions as the functional run - does that become sub-XX_ses-BL_field.nii.gz? And then the topup_results_fieldcoef.nii.gz becomes sub-XX_ses-BL_magnitude.nii.gz? And then you put the functional runs as the Intended for files in the json? And then add the readout time? And this would be a
Apologies if this is a daft question - I’m not familiar with the topup outputs and how they’re typically used.
Thanks so much!!
Hi,
Thanks so much Steven, you’re right, I should have been able to follow this. I was just a bit surprised that the undistorted image works as a field map but I guess that’s because it’s before topup has been applied so its more like the reverse phase, rather than a corrected image?
Thank you,
Maz