Hi Sean,
Apologies for the late reply.
After installing the master branch, and running the example script you wrote, the pipeline runs well until eddy.
I made the wrong assumption that I could use eddy_openmp, we cannot use cuda as we have a Radeon PRO WX 2100. Running eddy_openmp with the same inputs as the pipeline, I get EddyInputError: Slice-to-vol is not yet implemented for the CPU version. It looks like we will need GPU support ( MRtrix3 issue similar FSL issue). So the only workaround is buy an NVIDIA GPU or modify the eddy command so that it can run on the CPU. I may look into how I can still use eddy but without slice-to-volume correction, however, that seemed like one of the key benefits; to correct for intra slice movement.
For the moment, I have passed the use_mcflirt = True at the start of the file. And in
mcdc.mcdc(... do_dc=False, do_s2v=False, do_mbs=False, use_mcflirt=use_mcflirt,)
Unfortunate I cannot use eddy for the time being, but at least I can use more of the pipeline.
The next issue I had was in FIX. When the FIX directory is set up, the mask.nii.gz seems to be created; 1. By taking the T2 mask
2. Warping it into fMRI space
3. Binarising it
4. Thresholding out lowest 5% of values.
Since the original T2 mask I provided was already binarised, after the thresholding, the image turned to all zeros.
I also had a hard time finding the trained data using the pipeline, so I manually called fix -f and fix -c, to iron out these issues I inelegantly:
- Commented out line 136 in func/denoise.py
- Commented out lines 146-149 in func/denoise.py
- Commented out lines 304-306 in examples/ dhcp-dual-echo-time-fieldmap.py
- Then I removed the mask, added another one from earlier in the pipeline and ran fix extract and classify with some ugly code which I added in at line 303 in examples/ dhcp-dual-echo-time-fieldmap.py
fixDir = f"/home/johann.drayne/fmridhcpnew/output/sub-{subid}/ses-session1/denoise/"
textinFile = f"/home/johann.drayne/fmridhcpnew/output/sub-{subid}/ses-session1/denoise/fix4melview_train35_thr5.txt "
textoutFile = f"/home/johann.drayne/fmridhcpnew/output/sub-{subid}/ses-session1/denoise/fix_labels.txt"
bashCmd1 = "rm -r /home/johann.drayne/fmridhcpnew/output/sub-MS040053/ses-session1/denoise/mask.nii.gz"
bashCmd2 = "cp /home/johann.drayne/fmridhcpnew/output/sub-MS040053/ses-session1/mcdc/func_mcdc_brainmask.nii.gz /home/johann.drayne/fmr>
bashCmd3 = "/usr/local/fix/fix -f " + fixDir
bashCmd4 = "/usr/local/fix/fix -c " + fixDir + " /home/johann.drayne/dhcpfmri/data/train35.RData 5"
bashCmd5 = "mv " + textinFile + textoutFile
process = subprocess.Popen(bashCmd1.split(), stdout=subprocess.PIPE)
output, error = process.communicate()
print(output)
process = subprocess.Popen(bashCmd2.split(), stdout=subprocess.PIPE)
output, error = process.communicate()
print(output)
process = subprocess.Popen(bashCmd3.split(), stdout=subprocess.PIPE)
output, error = process.communicate()
print(output)
process = subprocess.Popen(bashCmd4.split(), stdout=subprocess.PIPE)
output, error = process.communicate()
print(output)
process = subprocess.Popen(bashCmd5.split(), stdout=subprocess.PIPE)
output, error = process.communicate()
print(output)
That ran FIX well, however there many of the ICA maps are unclassified so we are going to create our own training file and will update if there is much difference.
We had an issue in the STANDARD section with the pipeline not being able to find the atlas tree.
So we:
- Commented out line 320 in examples/ dhcp-dual-echo-time-fieldmap.py
- Added underneath
atlas_tree = Path('/home/johann.drayne/dhcpfmri/data/dhcp_volumetric_atlas_extended/atlas/atlas.tree')
We again faced a similar issue in with QC
- We did the same on line 650 in func/pipeline.py as above
- Similarly on line 500 in func/pipeline.py we changed
group_map = Path(util.get_setting('dhcp_group_maps'))
→
group_map = Path(‘/home/johann.drayne/dhcpfmri/data/group_maps.nii.gz’)
This brings me to the most recent error which I am struggling with.
The function I have gotten to in the main .py file is p.qc() on line 369.
The last function that the logger outputs is Add func_raw to QC standard this seems to be the function on line 702 in func/pipeline.py that is causing the issue. I have uploaded the last error output texterror.txt (6.2 KB)
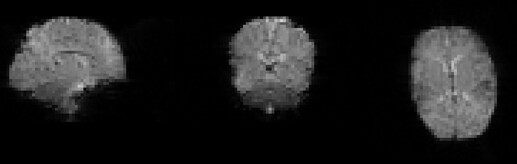
It is true qc/raw_dr2.nii.gz is not created, however, there are a couple things that suggest something else is at play here.
-
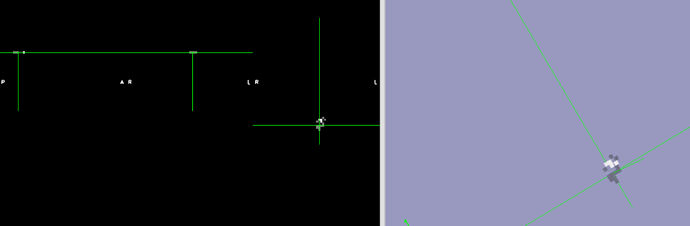
The output qc/raw_dseg.nii.gz is almost a fully empty image (). To me this suggests there is something going on with the warp file reg/func-mcdc_to_standard/func-mcdc_to_standard_invwarp.nii.gz
-
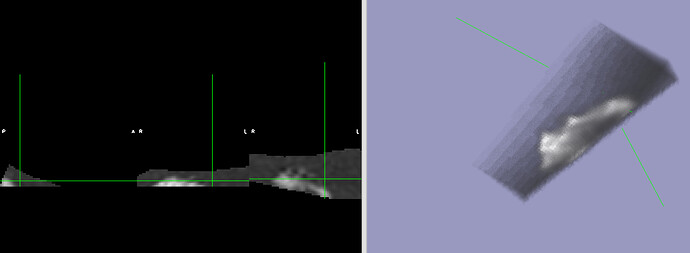
The output qc/raw_standard_tsnr.nii.gz seems to have been warped, however it is weirdly clipped. Maybe suggesting the qc/raw_tsnr.nii.gz was not aligned with the warped file?
-
The output qc/tmpkmepy1sl/template_native.nii.gz seems to be a purely grey image. I think this is to do with the warp reg/func-mcdc_to_standard/func-mcdc_to_standard_invwarp.nii.gz again.
-
The output qc/raw_dr1.txt is a text file of nans’ but I think this is because of the previous reasons.
-
Because dr1.txt is full of nans’ this is probably stopping qc/raw_dr2.nii.gz from being created.
To conclude, do you know a fix to run eddy, but only the CPU version?
and
Do you have any ideas as to how to combat this last issue? I will create our own training file and will also try the pipeline with another subject and see if that brings around any differences.
I would like to thank you for all your previous help so far, this pipeline is very exciting!
All the best,
Johann