Hi Anna,
To be clear, it’s not resampling, it’s reorienting, i.e., rotating the data matrix in order to flip the second and third axes. So, for a value that was indexed as datablock[i, j, k] will now be found at datablock[i, k, j]. Similarly, if we rincrement j, we’re moving in the inferior-superior direction: datablock_rsa[i, j + 1, k] = datablock_ras[i, k, j + 1].
This needs to be the same difference in “real-world” space (RAS+), so incrementing j by 1 moves ±4mm in the superior-inferior axis. Therefore, to keep the interpretation consistent, the voxel sizes (zooms) need to rotate to correspond to the new axes.
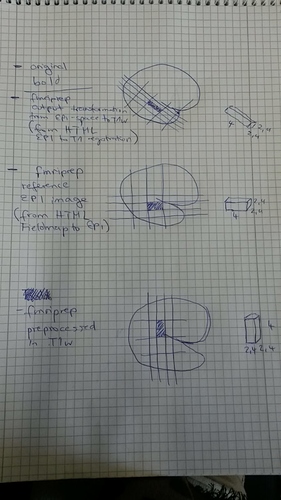
Just to think through the consequences, imagine that the voxel sizes stay the same while we rotate the data matrix. Suppose we have an image with (240mm)^3 field of view, in RSA orientation, with zooms 2.4mm, 4mm and 2.4mm. Then we have a data block of 100x60x100 voxels. If we rotate to RAS orientation, then our data block is now 100x100x60 voxels. If we don’t change the zooms, the new field of view will 240mm x 400mm x 144mm, so we’ll get stretching along the anterior/posterior dimension (remember, the voxel values have not changed), and compression along the inferior/superior dimension.
The obvious question then is why reorient at all. And the reason for that is that not all tools respect orientation information properly; as fMRIPrep uses tools from across a variety of software packages, keeping orientation consistent across all data that passes through fMRIPrep drastically reduces sources of variation; we validate that the pipeline works on a wide variety of RAS-oriented data, and then ensure that we feed the pipeline RAS-oriented data.
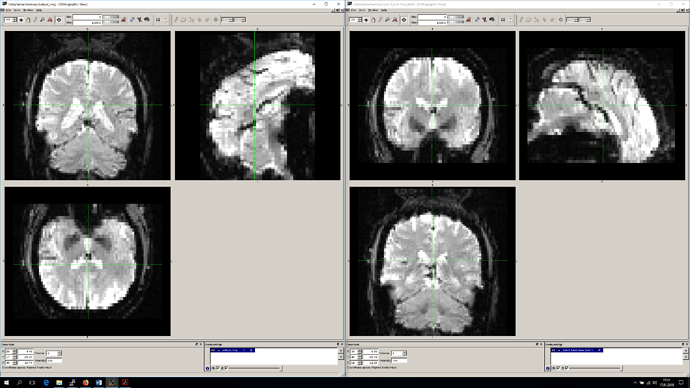
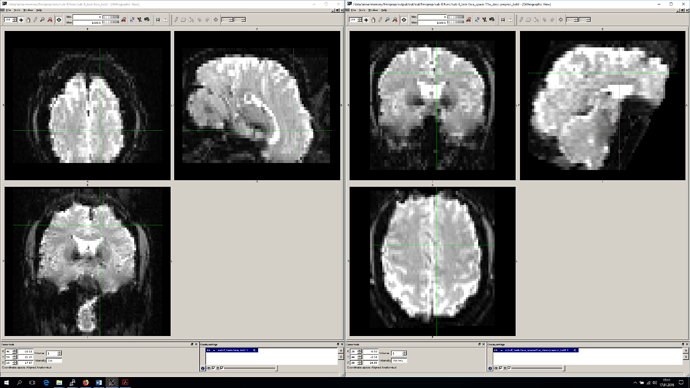
From what you’ve said, there isn’t a particular reason to keep the data in RSA orientation, just that you’d like to avoid resampling. I assure you that reorientation does not involve resampling. If you would like to check this out, you can use fslreorient2std or nibabel.as_closest_canonical on your original images, to verify that the data is unaffected:
fslreorient2std input_img.nii.gz output_img.nii.gz
import nibabel as nb
img = nb.load('input_img.nii.gz')
nb.as_closest_canonical(img).to_filename('output_img.nii.gz')
I believe fslreorient2std orients to either LAS or LPS, while nibabel will target RAS, but you can then take either of these outputs and overlay them with your original data in a viewer that respects orientation (fsleyes or freeview do, I know), and compare the appearance. If any resampling occurred, there should be a visible difference, but you should not see it.