Dear Neurostars,
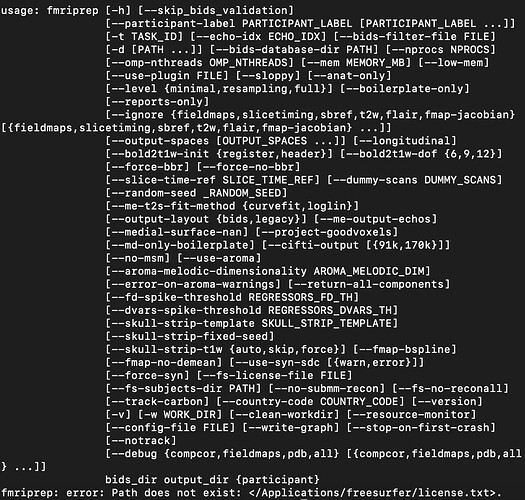
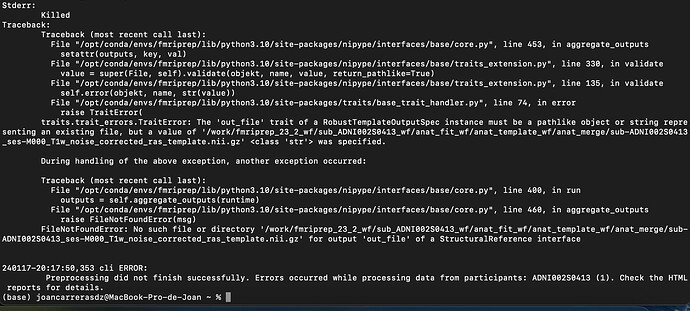
I have been trying to run fmriprep on a BIDS converted subject. However, when I apply the function “fmriprep BIDS_DIRECTORY DATA_PREP participant” in my terminal, appears the following message:
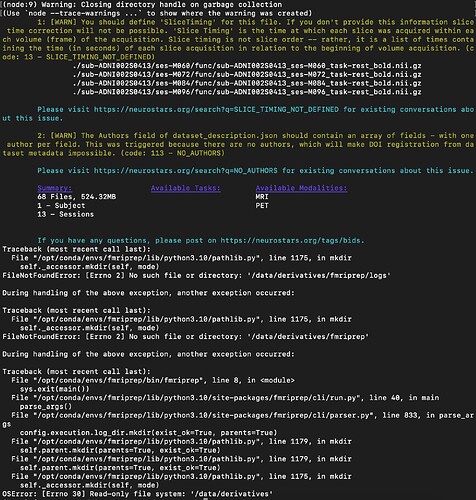
bids-validator does not appear to be installed
Traceback (most recent call last):
File "/Users/joancarrerasdiaz/miniconda3/bin/fmriprep", line 8, in <module>
sys.exit(main())
^^^^^^
File "/Users/joancarrerasdiaz/miniconda3/lib/python3.11/site-packages/fmriprep/cli/run.py", line 87, in main
setup_migas(init_ping=True)
File "/Users/joancarrerasdiaz/miniconda3/lib/python3.11/site-packages/fmriprep/utils/telemetry.py", line 200, in setup_migas
send_breadcrumb(status='R', status_desc='workflow start')
File "/Users/joancarrerasdiaz/miniconda3/lib/python3.11/site-packages/fmriprep/utils/telemetry.py", line 207, in send_breadcrumb
res = migas.add_project("nipreps/fmriprep", __version__, **kwargs)
^^^^^^^^^^^^^^^^^
AttributeError: module 'migas' has no attribute 'add_project'
Sentry is attempting to send 2 pending events
Waiting up to 2 seconds
Press Ctrl-C to quit
I have downloaded the bids-validator and reseted the terminal a couple of times and it keeps appearing the same message.
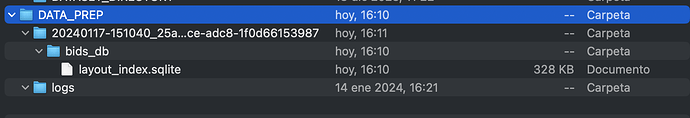
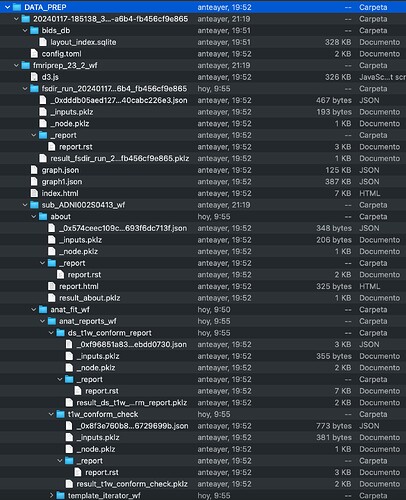
On the other hand, the data generated in the folder DATA_PREP is just a folder named logs, as you can see below.
/Users/joancarrerasdiaz/Desktop/CARPETES/UdG/Pràctiques/DATA_PREP
└── logs
How can I solve this problem?
Note: the structure of the folder
BIDS_DIRECTORY is the following one:
├── README
├── conversion_info
│ └── v0
│ ├── 11CPIB_pet_paths.tsv
│ ├── 18FAV1451_pet_paths.tsv
│ ├── 18FFDG_pet_paths.tsv
│ ├── amyloid_pet_paths.tsv
│ ├── dwi_paths.tsv
│ ├── flair_paths.tsv
│ ├── fmri_paths.tsv
│ └── t1_paths.tsv
├── dataset_description.json
├── participants.tsv
└── sub-ADNI002S0413
├── ses-M060
│ ├── func
│ │ ├── sub-ADNI002S0413_ses-M060_task-rest_bold.json
│ │ └── sub-ADNI002S0413_ses-M060_task-rest_bold.nii.gz
│ ├── pet
│ │ ├── sub-ADNI002S0413_ses-M060_trc-18FAV45_pet.nii.gz
│ │ ├── sub-ADNI002S0413_ses-M060_trc-18FFDG_rec-coregavg_pet.nii.gz
│ │ └── sub-ADNI002S0413_ses-M060_trc-18FFDG_rec-coregiso_pet.nii.gz
│ └── sub-ADNI002S0413_ses-M060_scans.tsv
├── ses-M072
│ ├── func
│ │ ├── sub-ADNI002S0413_ses-M072_task-rest_bold.json
│ │ └── sub-ADNI002S0413_ses-M072_task-rest_bold.nii.gz
│ └── sub-ADNI002S0413_ses-M072_scans.tsv
├── ses-M084
│ ├── func
│ │ ├── sub-ADNI002S0413_ses-M084_task-rest_bold.json
│ │ └── sub-ADNI002S0413_ses-M084_task-rest_bold.nii.gz
│ ├── pet
│ │ ├── sub-ADNI002S0413_ses-M084_trc-18FAV45_pet.nii.gz
│ │ ├── sub-ADNI002S0413_ses-M084_trc-18FFDG_rec-coregavg_pet.nii.gz
│ │ └── sub-ADNI002S0413_ses-M084_trc-18FFDG_rec-coregiso_pet.nii.gz
│ └── sub-ADNI002S0413_ses-M084_scans.tsv
├── ses-M096
│ ├── func
│ │ ├── sub-ADNI002S0413_ses-M096_task-rest_bold.json
│ │ └── sub-ADNI002S0413_ses-M096_task-rest_bold.nii.gz
│ └── sub-ADNI002S0413_ses-M096_scans.tsv
├── ses-M108
│ ├── func
│ ├── pet
│ │ └── sub-ADNI002S0413_ses-M108_trc-18FAV45_pet.nii.gz
│ └── sub-ADNI002S0413_ses-M108_scans.tsv
├── ses-M132
│ ├── dwi
│ │ ├── sub-ADNI002S0413_ses-M132_dwi.bval
│ │ ├── sub-ADNI002S0413_ses-M132_dwi.bvec
│ │ ├── sub-ADNI002S0413_ses-M132_dwi.json
│ │ └── sub-ADNI002S0413_ses-M132_dwi.nii.gz
│ ├── pet
│ │ ├── sub-ADNI002S0413_ses-M132_trc-18FAV1451_pet.nii.gz
│ │ └── sub-ADNI002S0413_ses-M132_trc-18FAV45_pet.nii.gz
│ └── sub-ADNI002S0413_ses-M132_scans.tsv
├── ses-M162
│ ├── pet
│ │ └── sub-ADNI002S0413_ses-M162_trc-18FAV45_pet.nii.gz
│ └── sub-ADNI002S0413_ses-M162_scans.tsv
└── sub-ADNI002S0413_sessions.tsv
Thank you,
Joan