Thank you for making such template available! However, I just tried it today and it ran into the error without no real output at all in the fmriprep folder. Also shell into the 1.4.1rc4 simg, it doesn’t show the regular templateflow folder as in 1.4.0.simg but only ICA-AROMA and freesurfer. Is there something wrong in my built singularity image?
Thanks!
@oesteban FYI I pulled the image through singularity, is the templateflow folder in a different place in fmriprep-1.4.1rc4 vs fmriprep-1.4.0? Thanks a lot for your help!
Correct, we rolled it back to $HOME/.cache/templateflow to make it easier for singularity users to set up.
If your singularity installation automatically binds $HOME then, it should work out of the box (although fMRIPrep will need to run at least once to initialize the templateflow home directory).
You can also overwrite the destination and bind the new directory. For instance, you can do:
export SINGULARITYENV_TEMPLATEFLOW_HOME=/opt/templateflow
singularity run -B /scratch/$USER/templateflow:/opt/templateflow ...
Thank you @oesteban! Can you confirm then the syntax below of using MNIPediatricAsym template is correct? I think that might be the reason why fMRIprep failed for me even at the anat processing level. As it was working for me before for the 1.4.0 version except changing the following lines:
–skull-strip-template MNIPediatricAsym:cohort-1 --output-spaces MNIPediatricAsym:cohort-1:res-2 anat fsaverage:den-10k
Yes, that command line should work. What is the error?
It throws the following error (and no out_pt141 folder being created yet at this point)
[Node] Finished “fmriprep_wf.single_subject_POLER01_wf.func_preproc_task_ADT_wf.bold_surf_wf.update_metadata”.
190702-17:57:24,128 nipype.workflow ERROR:
could not run node: fmriprep_wf.single_subject_POLER01_wf.anat_preproc_wf.anat_norm_wf.registration.a1
fMRIPrep failed: Workflow did not execute cleanly. Check log for details
Preprocessing did not finish successfully. Errors occurred while processing data from participants: POLER01 (3). Check the HTML reports for details
Hi @oesteban,
I downloaded templateflow to my local space /imaging/jj02/CALM/templateflow and have been trying to use the fix you suggested but I have a path error:
Failed to get real path of /var/singularity/mnt/final/imaging/jj02/CALM/templateflow: No such file or directory
There’s probably something simple I am missing here without understanding singularity very well, would be able to advise? Note I’m running this in Jupyter Lab and submitting to a HPC using SLURM.
Thank you.
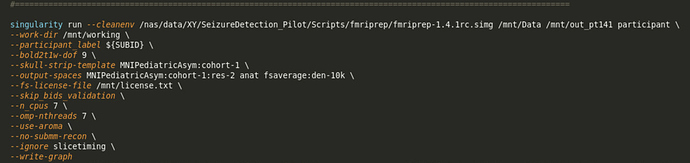
fmriprep1.4.1rc-MNIPediatric.txt (1.4 KB)
Hi @Xiaozhen_You,
We will need more information. Perhaps you can run fmriprep adding the flag -vv for increased verbosity and attach the full log of execution.
Additionally, you should find what we call “crashfiles” under fmriprep/sub-${SUBID}/log/, please list those files for a subject and attach the least recent error.
BTW, it would also help if you used text instead of screen captures so we can copy and paste paths and other relevant pieces of information.
Hi @JoffJones,
I would recommend simplifying your execution workflow. IMHO there are too many layers of software for a single submission. Can you just run sbatch manually on a file like this?
#!/bin/bash
#
#SBATCH -J fmriprep
#SBATCH --array=1
#SBATCH --time=48:00:00
#SBATCH -n 1
#SBATCH --cpus-per-task=16
#SBATCH --mem-per-cpu=4G
#SBATCH -p normal
# Outputs ----------------------------------
#SBATCH -o log/%x-%A-%a.out
#SBATCH -e log/%x-%A-%a.err
#SBATCH --mail-user=%u@yourdomain.com
#SBATCH --mail-type=ALL
# ------------------------------------------
BIDS_DIR="/imaging/projects/cbu/calm/CALM_BIDS_New"
OUTPUT_DIR="${BIDS_DIR}/derivatives/fmriprep-1.4.1rc4"
WORK_DIR="${BIDS_DIR}/derivatives/fmriprep-1.4.1rc4-work"
TEMPLATEFLOW_DIR="/scratch/$USER/templateflow"
SINGULARITY_CMD="singularity run --cleanenv -B ${BIDS_DIR}:/data -B ${OUTPUT_DIR}:/out -B ${WORK_DIR}:/work -B ${TEMPLATEFLOW_DIR}:/opt/templateflow /imaging/local/software/singularity_images/fmriprep/fmriprep-1.4.1rc4.simg"
export SINGULARITYENV_FS_LICENSE=$HOME/.freesurfer.txt
export SINGULARITYENV_TEMPLATEFLOW_HOME=/opt/templateflow
subject=$( printf "%03d" $SLURM_ARRAY_TASK_ID )
cmd="${SINGULARITY_CMD} /data /out participant --participant-label $subject -w /work --skull-strip-template MNIPediatricAsym:cohort-1 --output-spaces MNIPediatricAsym:cohort-1:res-2 --fs-no-reconall --omp-nthreads 8 --nthreads 12 --mem_mb 30000 --notrack -vv"
echo Running task ${SLURM_ARRAY_TASK_ID}
echo Commandline: $cmd
eval $cmd
exitcode=$?
echo "sub-$subject ${SLURM_ARRAY_TASK_ID} $exitcode" \
>> ${SLURM_JOB_NAME}.${SLURM_ARRAY_JOB_ID}.tsv
echo Finished tasks ${SLURM_ARRAY_TASK_ID} with exit code $exitcode
exit $exitcode
Please do not forget to adjust the SBATCH arguments to something that fits your settings. In this regard, I identified that you were requesting 64GB per cpu. I don’t know whether you were planning to run fMRIPrep single-threaded, otherwise that is likely the reason SLURM didn’t even try to run it.
Please report all the output you get from every command you run.
Thanks
Hi @oesteban,
Thank you for this! I modified the bash script to point to a valid freesurfer license and added +1 to the subject so it begins at ‘sub-001’. It also couldn’t find templateflow, but I redirected to a version that I downloaded - the only problem is that the nifti files are only 1kb and there are a number of itk errors when trying to read them. Is templateflow meant to contain the actual T1w template?
#!/bin/sh
#
#SBATCH -J fmriprep
#SBATCH --array=1
#SBATCH --time=48:00:00
#SBATCH -n 1
#SBATCH --cpus-per-task=16
#SBATCH --mem-per-cpu=4G
#SBATCH -p normal
# Outputs ----------------------------------
#SBATCH -o log/%x-%A-%a.out
#SBATCH -e log/%x-%A-%a.err
#SBATCH --mail-user=%u@yourdomain.com
#SBATCH --mail-type=ALL
# ------------------------------------------
BIDS_DIR="/imaging/jj02/CALM/bids_test2"
OUTPUT_DIR="${BIDS_DIR}/derivatives/fmriprep-1.4.1rc4"
WORK_DIR="${BIDS_DIR}/derivatives/fmriprep-1.4.1rc4-work"
TEMPLATEFLOW_DIR="/imaging/jj02/CALM/templateflow"
CALM_DIR="/imaging/jj02/CALM"
SINGULARITY_CMD="singularity run --cleanenv -B ${BIDS_DIR}:/data -B ${OUTPUT_DIR}:/out -B ${WORK_DIR}:/work -B ${TEMPLATEFLOW_DIR}:/opt/templateflow -B ${CALM_DIR}:/CALM /imaging/local/software/singularity_images/fmriprep/fmriprep-1.4.1rc4.simg"
export SINGULARITYENV_FS_LICENSE=/imaging/jj02/CALM/license.txt
export SINGULARITYENV_TEMPLATEFLOW_HOME=/opt/templateflow
subject=$( printf "%03d" $SLURM_ARRAY_TASK_ID +1)
cmd="${SINGULARITY_CMD} /data /out participant --participant-label $subject -w /work --skull-strip-template MNIPediatricAsym:cohort-1 --output-spaces MNIPediatricAsym:cohort-1:res-2 --fs-no-reconall --fs-license-file /CALM/license.txt --omp-nthreads 8 --nthreads 12 --mem_mb 30000 --notrack -vv"
echo Running task ${SLURM_ARRAY_TASK_ID}
echo Commandline: $cmd
eval $cmd
exitcode=$?
echo "sub-$subject ${SLURM_ARRAY_TASK_ID} $exitcode" \
>> ${SLURM_JOB_NAME}.${SLURM_ARRAY_JOB_ID}.tsv
echo Finished tasks ${SLURM_ARRAY_TASK_ID} with exit code $exitcode
exit $exitcodeI found the templateflow directory located in /$HOME/.cache/templateflow which seems to have the correct T1w templates. When I use this as my TEMPLATE_FLOW_DIR I get a SSL connection error (see below). This might be related to our firewall, is it necessary for fmriprep to establish a web connection?
Commandline: singularity run --cleanenv -B /imaging/jj02/CALM/bids_test2:/data -B /imaging/jj02/CALM/bids_test2/derivatives/fmriprep-1.4.1rc4:/out -B /imaging/jj02/CALM/bids_test2/derivatives/fmriprep-1.4.1rc4-work:/work -B //home/jj02/.cache/templateflow:/opt/templateflow -B /imaging/jj02/CALM:/CALM /imaging/local/software/singularity_images/fmriprep/fmriprep-1.4.1rc4.simg /data /out participant --participant-label 001 -w /work --skull-strip-template MNIPediatricAsym:cohort-1 --output-spaces MNIPediatricAsym:cohort-1:res-2 --fs-no-reconall --fs-license-file /CALM/license.txt --omp-nthreads 8 --nthreads 12 --mem_mb 30000 --notrack -vv
Making sure the input data is BIDS compliant (warnings can be ignored in most cases).
1: [WARN] The Authors field of dataset_description.json should contain an array of fields - with one author per field. Thiswas triggered based on the presence of only one author field. Please ignore if all contributors are already properly listed. (code:102 - TOO_FEW_AUTHORS)
Please visit https://neurostars.org/search?q=TOO_FEW_AUTHORS for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
56 Files, 388.54MB rest T1w
5 - Subjects T2w
1 - Session dwi
bold
If you have any questions, please post on https://neurostars.org/tags/bids.
190704-11:13:49,24 nipype.workflow IMPORTANT:
Running fMRIPREP version 1.4.1rc4:
* BIDS dataset path: /data.
* Participant list: ['001'].
* Run identifier: 20190704-111349_03bdc74f-c991-4d6c-af9c-89616cde09bf.
190704-11:13:50,502 nipype.workflow IMPORTANT:
Creating bold processing workflow for "/data/sub-001/func/sub-001_task-rest_bold.nii.gz" (0.03 GB / 270 TRs). Memory resampled/largemem=0.13/0.21 GB.
190704-11:13:50,504 nipype.workflow IMPORTANT:
No single-band-reference found for sub-001_task-rest_bold.nii.gz
Downloading https://templateflow.s3.amazonaws.com/tpl-MNI152NLin2009cAsym/tpl-MNI152NLin2009cAsym_res-02_desc-fMRIPrep_boldref.nii.gz
Process Process-2:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/contrib/pyopenssl.py", line 444, in wrap_socket
cnx.do_handshake()
File "/usr/local/miniconda/lib/python3.7/site-packages/OpenSSL/SSL.py", line 1907, in do_handshake
self._raise_ssl_error(self._ssl, result)
File "/usr/local/miniconda/lib/python3.7/site-packages/OpenSSL/SSL.py", line 1639, in _raise_ssl_error
_raise_current_error()
File "/usr/local/miniconda/lib/python3.7/site-packages/OpenSSL/_util.py", line 54, in exception_from_error_queue
raise exception_type(errors)
OpenSSL.SSL.Error: [('SSL routines', 'tls_process_server_certificate', 'certificate verify failed')]
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/connectionpool.py", line 600, in urlopen
chunked=chunked)
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/connectionpool.py", line 343, in _make_request
self._validate_conn(conn)
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/connectionpool.py", line 849, in _validate_conn
conn.connect()
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/connection.py", line 356, in connect
ssl_context=context)
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/util/ssl_.py", line 359, in ssl_wrap_socket
return context.wrap_socket(sock, server_hostname=server_hostname)
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/contrib/pyopenssl.py", line 450, in wrap_socket
raise ssl.SSLError('bad handshake: %r' % e)
ssl.SSLError: ("bad handshake: Error([('SSL routines', 'tls_process_server_certificate', 'certificate verify failed')])",)
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/site-packages/requests/adapters.py", line 445, in send
timeout=timeout
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/connectionpool.py", line 638, in urlopen
_stacktrace=sys.exc_info()[2])
File "/usr/local/miniconda/lib/python3.7/site-packages/urllib3/util/retry.py", line 398, in increment
raise MaxRetryError(_pool, url, error or ResponseError(cause))
urllib3.exceptions.MaxRetryError: HTTPSConnectionPool(host='templateflow.s3.amazonaws.com', port=443): Max retries exceeded with url: /tpl-MNI152NLin2009cAsym/tpl-MNI152NLin2009cAsym_res-02_desc-fMRIPrep_boldref.nii.gz (Caused by SSLError(SSLError("bad handshake:Error([('SSL routines', 'tls_process_server_certificate', 'certificate verify failed')])")))
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.7/multiprocessing/process.py", line 297, in _bootstrap
self.run()
File "/usr/local/miniconda/lib/python3.7/multiprocessing/process.py", line 99, in run
self._target(*self._args, **self._kwargs)
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/cli/run.py", line 608, in build_workflow
work_dir=str(work_dir),
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/base.py", line 259, in init_fmriprep_wf
use_syn=use_syn,
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/base.py", line 609, in init_single_subject_wf
use_syn=use_syn,
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/bold/base.py", line 459, in init_func_preproc_wf
bold_reference_wf = init_bold_reference_wf(omp_nthreads=omp_nthreads)
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/bold/util.py", line 124, in init_bold_reference_wf
omp_nthreads=omp_nthreads, pre_mask=pre_mask)
File "/usr/local/miniconda/lib/python3.7/site-packages/fmriprep/workflows/bold/util.py", line 291, in init_enhance_and_skullstrip_bold_wf
'MNI152NLin2009cAsym', resolution=2, desc='fMRIPrep', suffix='boldref')
File "/usr/local/miniconda/lib/python3.7/site-packages/templateflow/api.py", line 39, in get
_s3_get(filepath)
File "/usr/local/miniconda/lib/python3.7/site-packages/templateflow/api.py", line 130, in _s3_get
r = requests.get(url, stream=True)
File "/usr/local/miniconda/lib/python3.7/site-packages/requests/api.py", line 72, in get
return request('get', url, params=params, **kwargs)
File "/usr/local/miniconda/lib/python3.7/site-packages/requests/api.py", line 58, in request
return session.request(method=method, url=url, **kwargs)
File "/usr/local/miniconda/lib/python3.7/site-packages/requests/sessions.py", line 512, in request
resp = self.send(prep, **send_kwargs)
File "/usr/local/miniconda/lib/python3.7/site-packages/requests/sessions.py", line 622, in send
r = adapter.send(request, **kwargs)
File "/usr/local/miniconda/lib/python3.7/site-packages/requests/adapters.py", line 511, in send
raise SSLError(e, request=request)
requests.exceptions.SSLError: HTTPSConnectionPool(host='templateflow.s3.amazonaws.com', port=443): Max retries exceeded with url: /tpl-MNI152NLin2009cAsym/tpl-MNI152NLin2009cAsym_res-02_desc-fMRIPrep_boldref.nii.gz (Caused by SSLError(SSLError("bad handshake: Error([('SSL routines', 'tls_process_server_certificate', 'certificate verify failed')])")))
Finished tasks with exit code 1Thanks for your patience, that is a new error. Please allow me some time to identify the root of it - probably some missing dependency in the singularity image.
cc/ @effigies is there anything obvious that jumps to your eyes?
If it’s persistent, it’s likely a firewall issue. If it’s intermittent, then it may be a problem with Amazon’s CDN. I think the solution is likely to be to cache the template, whether in the Singularity image or on the host.
Thanks for your help, we managed to find a workaround by downloading the templateflow directory and referring to it in the singularity command. There was an issue with our firewall so we downloaded it from a http rather than https link and we also had to download an adult MNI template separately, as this was used later down the line as a bold reference image:
import templateflow.api
templateflow.api.TF_S3_ROOT = ‘http://templateflow.s3.amazonaws.com’
get(‘MNI152NLin2009cAsym’)
Then we were able to run this in our Jupyter Notebook as:
import os
my_env = os.environ.copy()
my_env[“SINGULARITYENV_TEMPLATEFLOW_HOME”] = “/templateflow”
for n in range(1):
p = subprocess.run(" ".join([“singularity”, “exec”, “-C”,
“-B”, “/imaging/jj02/CALM:/CALM”,
“-B”, “/imaging/projects/cbu/calm/CALM_BIDS_New:/CALM_BIDS_New”,
“-B”, “/home/jj02/.cache/templateflow:/templateflow”,
“/imaging/local/software/singularity_images/fmriprep/fmriprep-1.4.1rc4.simg”,
“fmriprep”,
“/CALM_BIDS_New”,
“/CALM_BIDS_New/derivatives/fmriprep_child_test6”,
“participant”, “–participant_label”, ‘%03d’ %(n+1),
“-v”, “-w”, “/CALM_BIDS_New/derivatives/fmriprepwork_child_test6”,
“–skull-strip-template”, “MNIPediatricAsym:cohort-1”,
“–output-spaces”, “MNIPediatricAsym:cohort-1”,
“–fs-no-reconall”,
“–fs-license-file”, “/CALM/license.txt”,
“–notrack”]),
shell=True, stdout=subprocess.PIPE, stderr=subprocess.PIPE, env=my_env)
print(p.args)
print(p.stdout.decode())
print(p.stderr.decode())
Would you like to contribute this workaround into the TemplateFlow client?
Hi, I have been helping Joff with this. We could certainly contribute something. Are you thinking about just modifying the get method to fall back on http if https fails? Happy to do this if you’re happy, but I’m not sure it’s great from a security standpoint.
We actually realised subsequently that this SSL error could have been avoided if we had just set the REQUESTS_CA_BUNDLE environment variable to point to our site-specific certificate (certifi checks this variable, so this takes care of all urllib3 SSL issues for us). I guess we could have passed this environment variable into the container instead of the bind-mount and the TEMPLATEFLOW_HOME variable. So in summary, the connection problems were probably more caused by our bad config than anything wrong with templateflow.
Although it would maybe be useful if the fmriprep container had a more obvious route for passing a custom templates directory. It seems a little inefficient to download templates every time you run the container.
It’d be good to have this in the documentation :). Thanks very much!
For singularity is actually necessary that you bind some writable place into TEMPLATEFLOW_HOME.
In version 1.4.1, TEMPLATEFLOW_HOME is not set. It can be set from outside the container with export SINGULARITYENV_TEMPLATEFLOW_HOME=/some/path/bound/from/host.
Since the env var is not set and most Singularity installations automatically bind $HOME, and reset $HOME inside the container, then TemplateFlow will write to $HOME/.cache/templateflow, which will be generally writable.