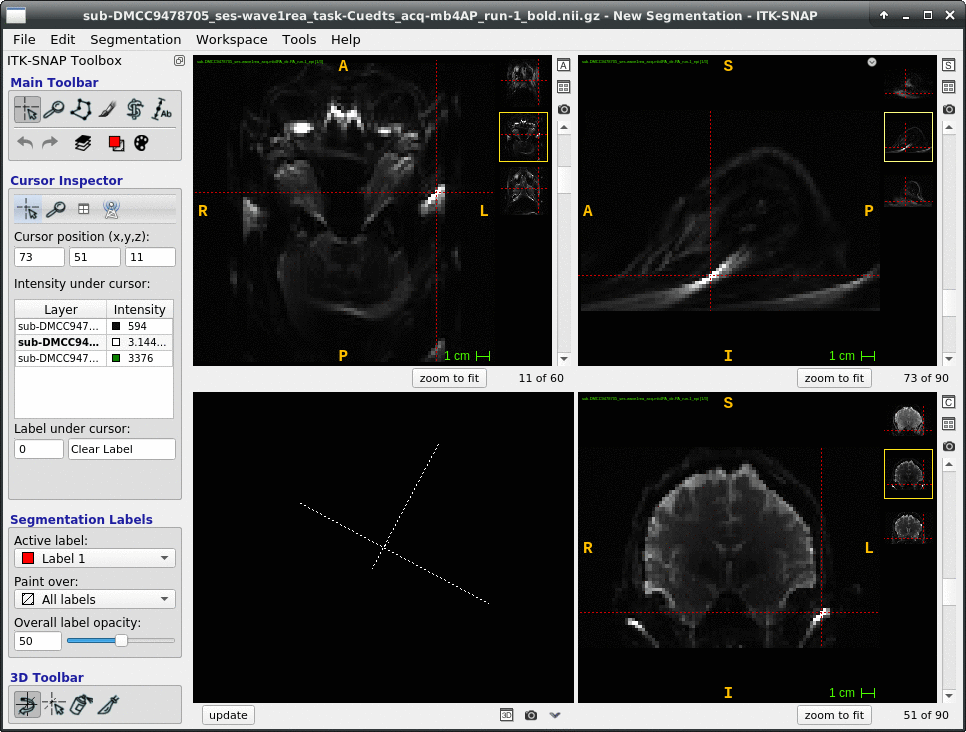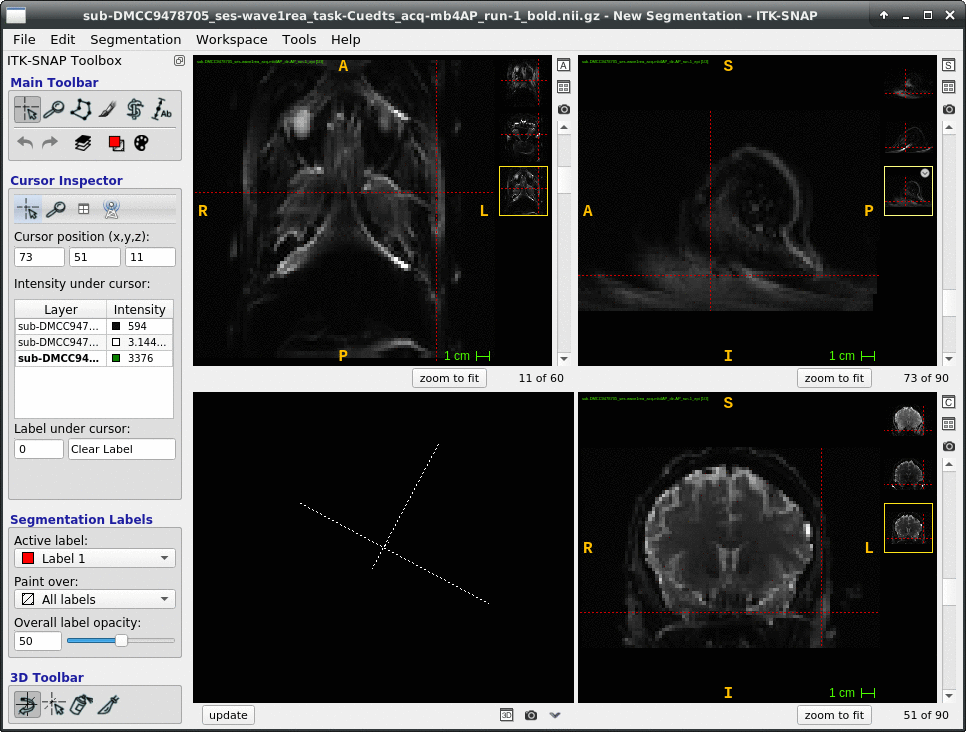
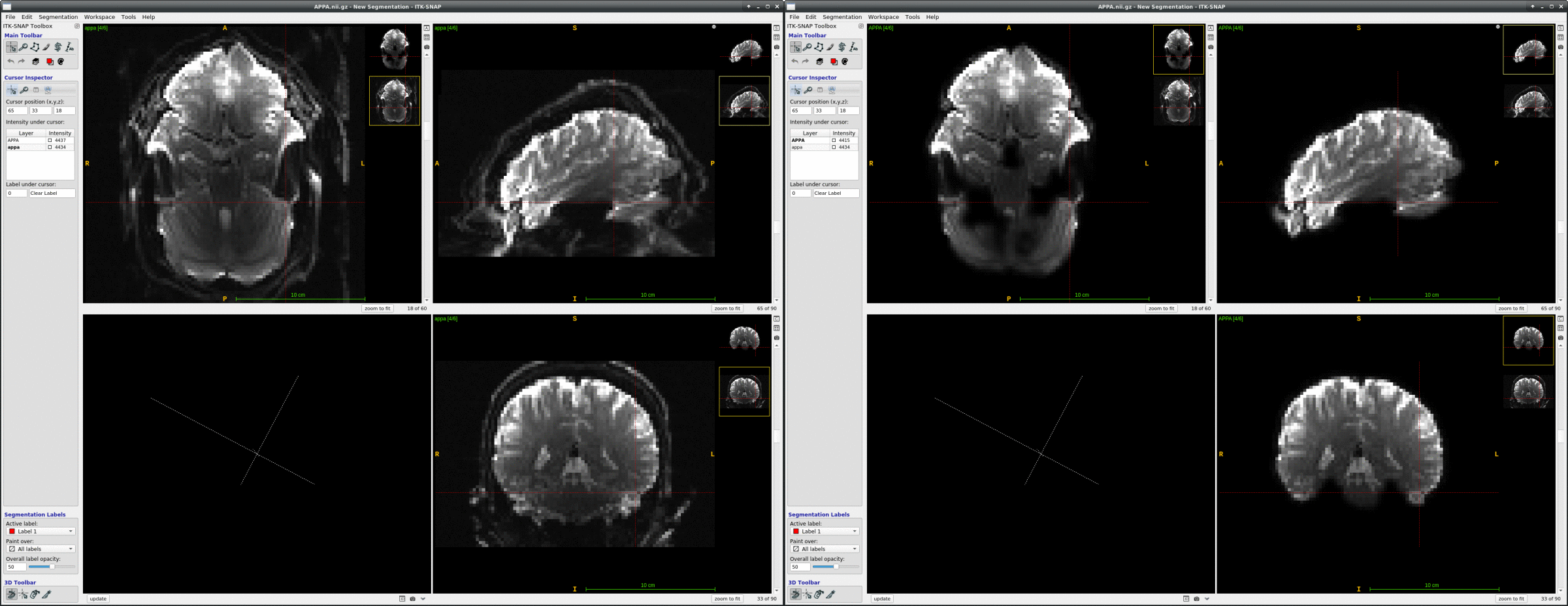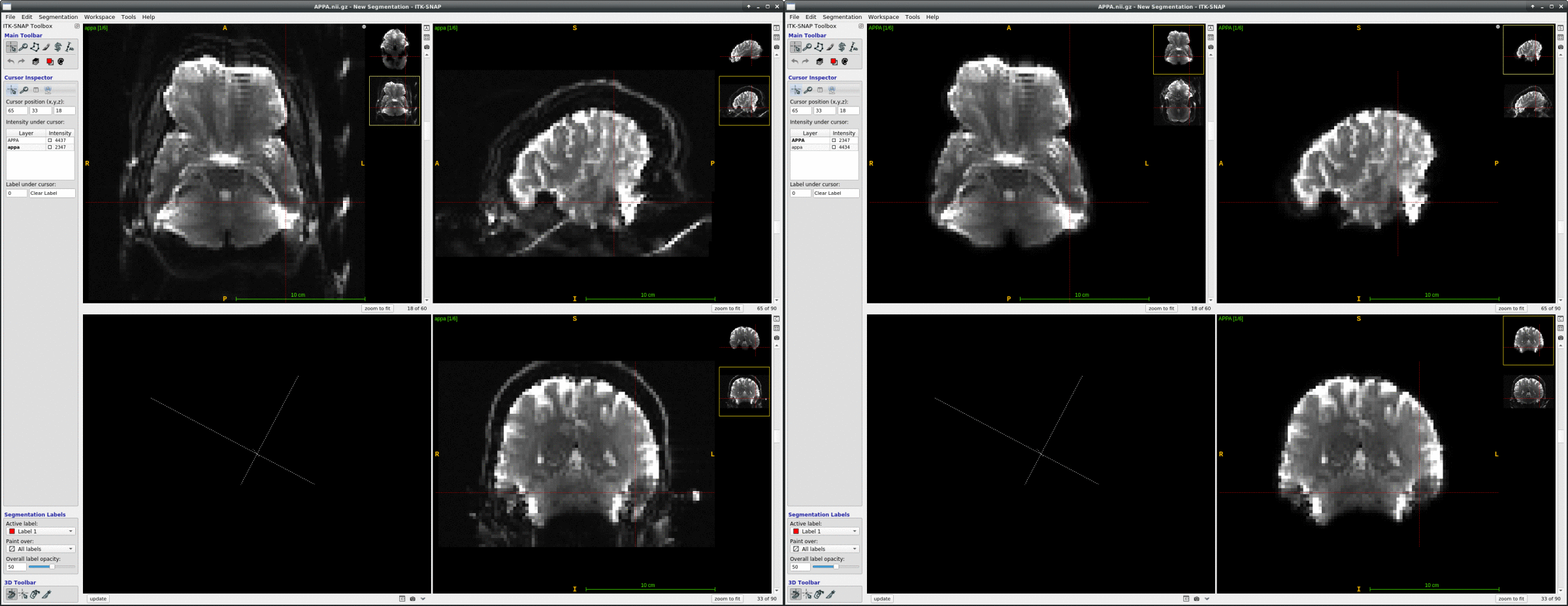
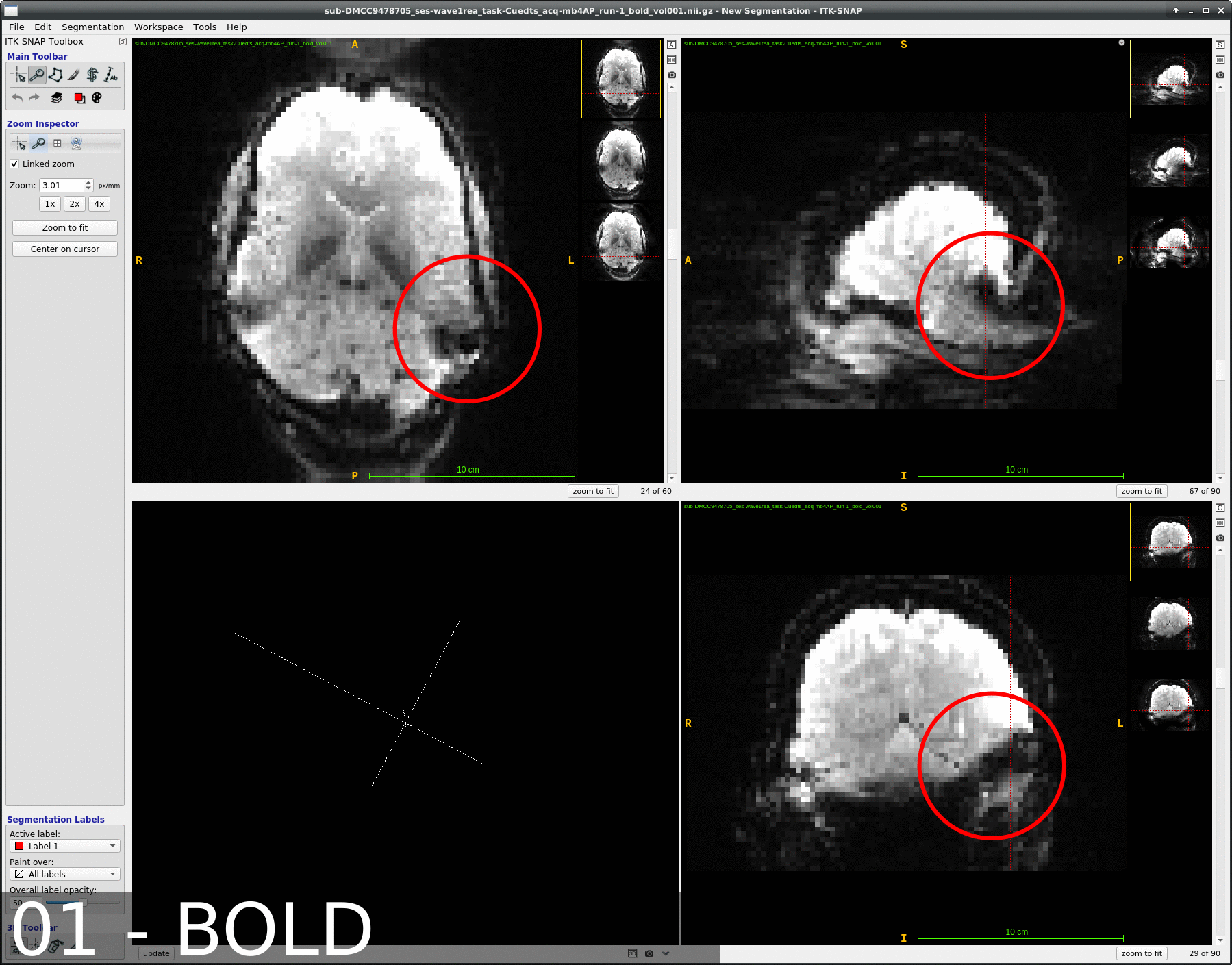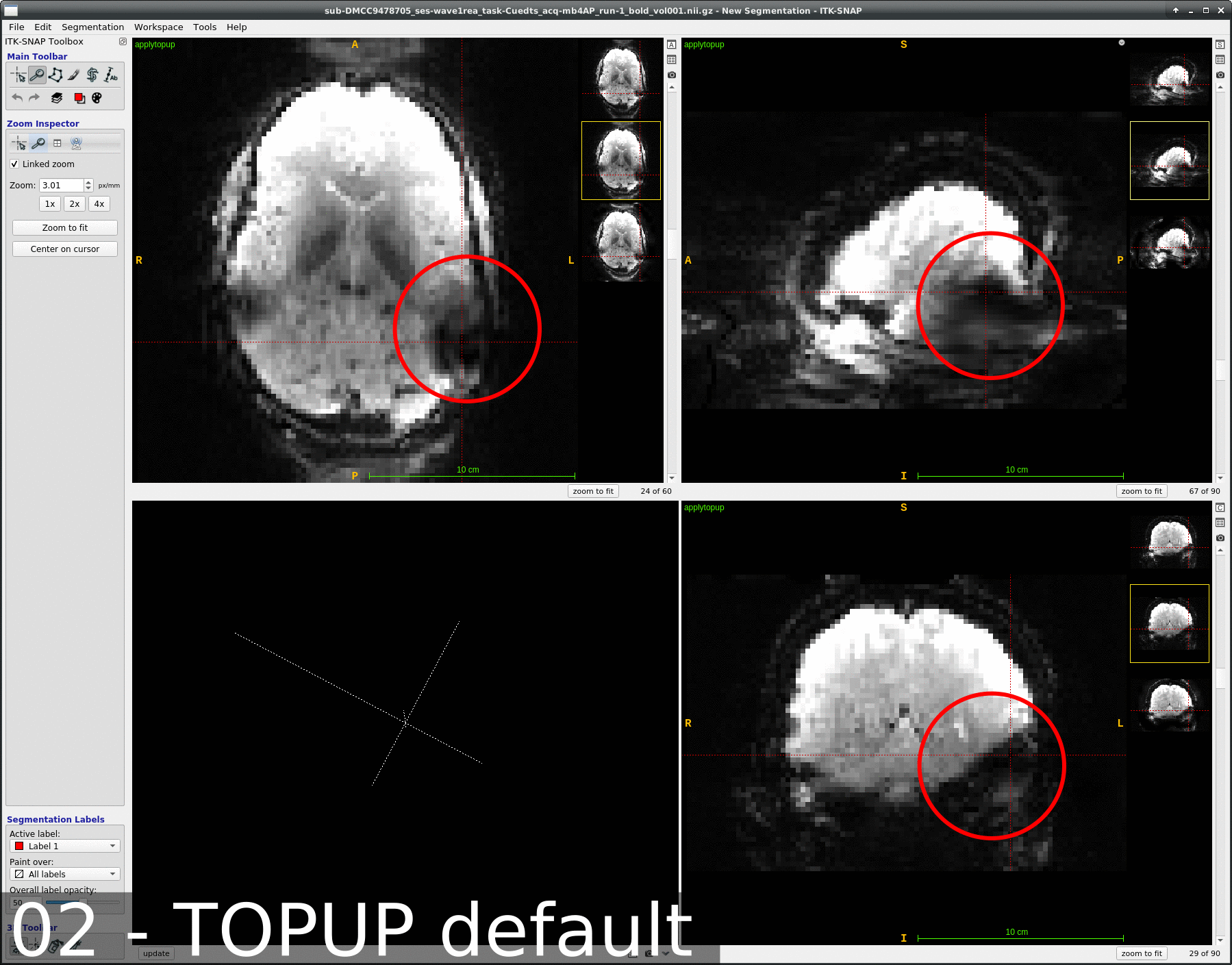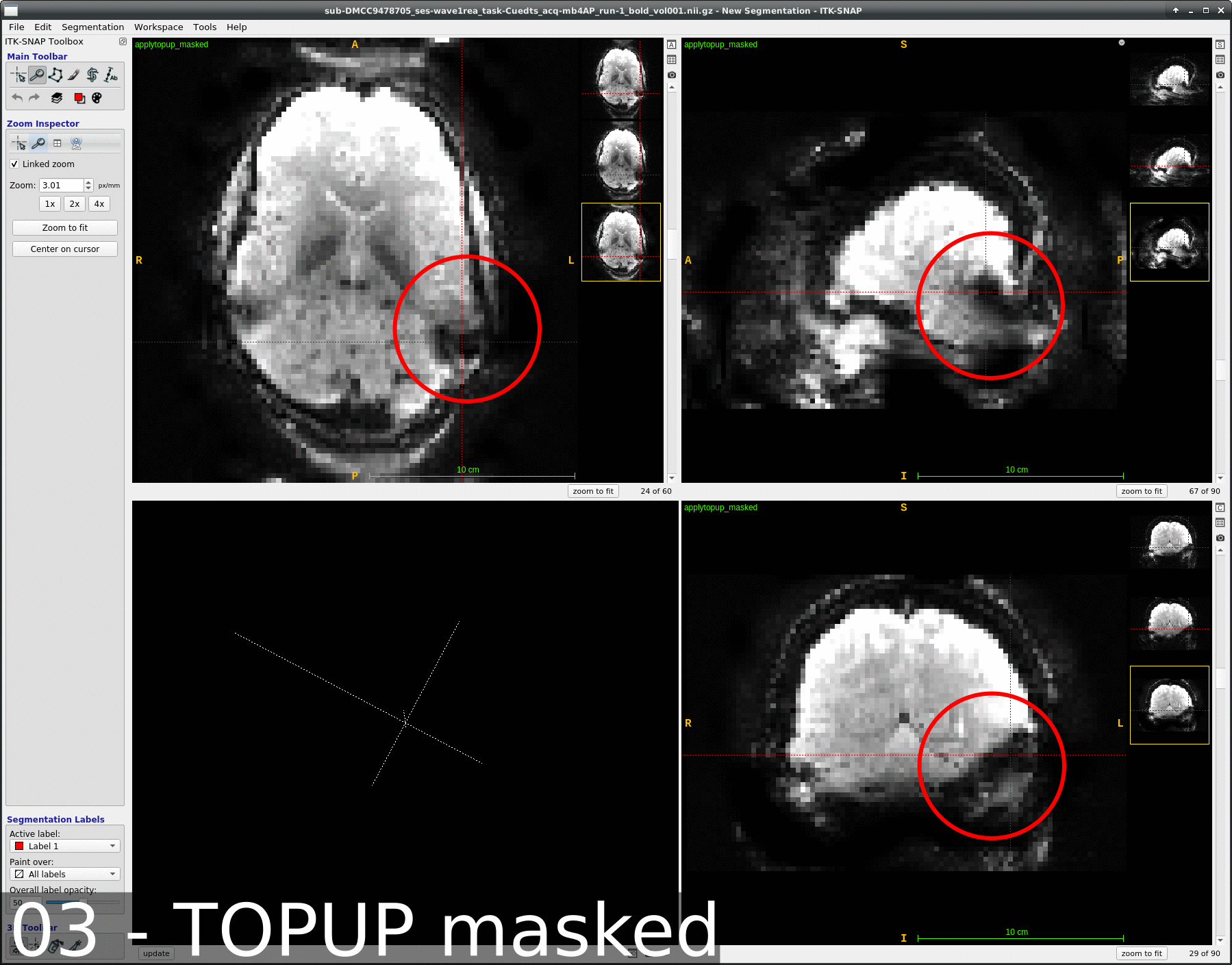
We occasionally have very strange fmriprep preprocessing output, such as the brain in the _space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz image being clearly not MNI-shaped or not centered. These errors often correct themselves when the preprocessing is repeated from the beginning (similar to this).
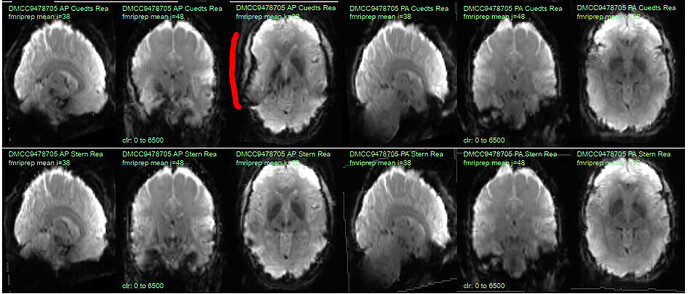
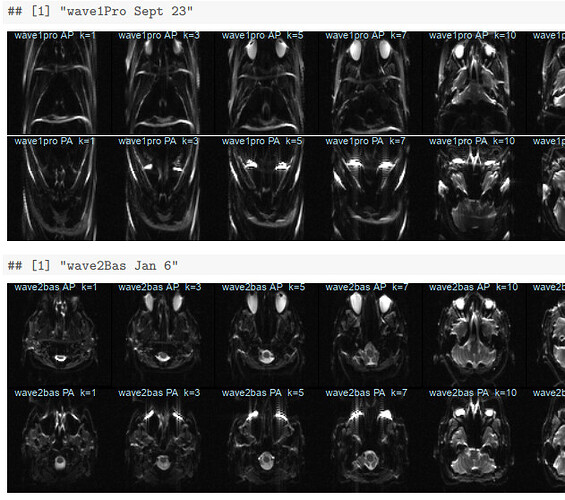
This “do it again until it works” strategy is failing for a particular session, however, and I am out of debugging ideas. Three runs in one session for a DMCC participant are affected, AP encoding runs only. Fieldmaps were collected at the beginning of the session, then two runs (AP then PA) of each task, in the order Stern, Stroop, Cuedts, Axcpt. The distortion increased over the course of the session; Stern seems fine, Stroop a bit affected, Cuedts and Axcpt worst. The participant did not leave the scanner during the session.
Here are temporal mean images to give an idea of the distortion; the lower row are the Stern runs (AP in first three columns, PA in second), the top Cuedts.
This person’s motion is quite typical (better than many); the dicoms and dcm2niix-converted functional niftis also look typical, as do the SBRef and fieldmap images. Our standard pipelines use older versions of the software; for debugging we converted the dataset using the newest version of dcm2niix and fmriprep 20.2.3, but without noticeable improvement.
Any suggestions for fixing this? Or ideas of what is going wrong? The last two AP runs of this session (Cuedts and Axcpt) are in the BIDS subdirectory of https://wustl.box.com/s/rp7wop16km39736r0thsq9o54h2klmhf, with accompanying anatomy*, SBRef, and fieldmap files. The fmriprep 20.2.3 output is in the fmriprep subdirectory, including the html summary file.
Thanks!
* for debugging we did not include defacing, so I removed the T1 and T2 images from the BIDS version; let me know if you need them.