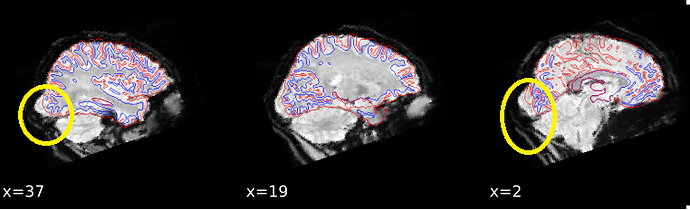
We are seeing some strange results of the susceptibility distortion correction in fmriprep v21.0.2. It appears to stretch the brain too far in many participants, making alignment with the structural data worse. The data has A>P phase encoding direction and we use phase difference fieldmaps to do SDC. We checked the settings for the phase encoding direction in the json files (it’s set to ‘j-’. We actually ran it with ‘j’ as well but the results of that were even worse). Earlier time points of this dataset have also been processed with an older version of fmriprep (1.5.2) and that did not result in any SDC problems and led to better alignment than these results from v21.0.2. Any ideas what is happening here?
Hi,
A few questions:
- does this happen in all, or just some participants?
- have you tried the fieldmap-less method?
- does this error persist in v22.0.0?
- what scanner manufacturer do the images come from? If GE, this discussion may be useful: fMRIPrep 20.2.3 directly measured B0 map SDC correction created worse functional images
- are there any errors to report?
- are you sure that no previous derivatives from the earlier run (e.g the work directory) were used in the newer run?
Best,
Steven
Thank you for your questions, Steven! Below are the answers:
- More clearly in some participants than others, but visible in the vast majority
- Have not tried this, but will look at it in the next couple days
- Have not tried this yet. Do you expect it to make a difference? I don’t see major changes to the SDC from 21.0.2 to 22.0. It looks like issues with SDC are still being worked out (“For the next release, we are investigating issues with susceptibility distortion correction (SDC).”)
- It’s a Siemens Skyra
- No errors
- We had already removed the working directories from the processing with an earlier version
I suggest looking at the field maps and/or single band reference images; abnormalities in those can lead to strangely distorted functional runs after preprocessing.
This topic has such a case: Repeated fmriprep distortion; perhaps from susceptibility distortion, and I summarized it and a few other examples at MVPA Meanderings: troubleshooting run-level failed preprocessing.
I aslo met the same problems. Many subjects have the unusual SDC and even I rerun the preprocessing. Some good results become the distorted reuslts. Did you solve the problem?
Same here, Yukun! Unfortunately, we still see problems when using fieldmapless correction; the SDC distortion doesn’t seem to depend on the quality of our fieldmaps. We also see normal results when processing the same scan with an older version of fmriprep so we plan to revert back to v20. However, I’d be really curious to hear if you come across a solution for this that works with the newer version of fmriprep.
Do you confirm that the SDC method used in all these problematic cases are with the Phase Difference Fieldmap method?
One could look at the implementation of this correction method in fmriprep v20 and in fmriprep v21 (i.e. in SDCFlows as SDC was excised from FMRIPREP at this point) to see if there is a difference in the implementation of the Phase difference Fieldmap method.
@cgw, on the image that is shown on the first post of this thread, the defect seems very localized in the occipital part of the brain. Could you show us how the estimated fieldmap looks like for this subject, calculated by fmriprep v21, and if you have it, by fmriprep v20?
Thanks!
Thanks. I will keep you posted if we make any progress.
Unfortunately, the fmriprep v22 seems have same problem.
I rerun the preprocessing with v22. I used 8 subjects to test and half of them have different bad correction. Even some of runs have good correction in the previous preprocessing.
I post some information in this link.
If you are interested in this question,I have show my problem in this link.